Content uploaded by Mohd Aamir
Author content
All content in this area was uploaded by Mohd Aamir on Jun 12, 2021
Content may be subject to copyright.
Nova Hedwigia, Vol. 109 (2019), Issue 1-2, 81–93 Article
Published online May 20, 2019; published in print August 2019
First report of Newbya recurva (Saprolegniaceae) from
India
Manish Kumar Dubey1*, Timothy Yong James2, Andleeb Zehra1,
Mohd. Aamir1 and Ram Sanmukh Upadhyay1
1 Laboratory of Mycopathology and Microbial Technology, Centre of Advanced Study in Botany,
Institute of Science, Banaras Hindu University, Varanasi-221005, Uttar Pradesh, India
2 Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109,
U.S.A.
* Corresponding author: mkmkdubey@gmail.com
With 2 figures
Abstract: In the present report, Newbya recurva (Cornu) Dick and Spencer is isolated, described,
illustrated and identified based on morphological characters and phylogenetic analysis of the ITS-
rDNA region. This formerly Achlya species is mainly recognized by the presence of achlyoid type
of zoospore discharge, spherical oogonia with external ornamented wall provided with numerous,
stout, symmetrically arranged conical truncate and thin-walled projections/ protrusions at the end
(autapomorphic or hallmark feature); large centric or subcentric oospores, which occasionally
failed to mature, generally ranging from 1–5 per oogonium, and predominantly androgynous an-
theridial branches. Apart from these morphological features, the identity of the specimens was
further confirmed by ITS-rDNA sequence comparison. The result of phylogenetic analysis sup-
ported the prevailing idea that Achlya recurva and N. recurva should be considered a single species
in the genus Newbya. A short description, comments, and illustration, accompanied with its com-
parison with other allied taxa of the genus as well as a molecular phylogeny of the ITS region are
provided in this paper. This study also confirms the first report of N. recurva based on morphologi-
cal as well as analysis of ITS-rDNA from India.
Key words: Chandra Prabha Wildlife Sanctuary, Water-mold, Oomycota, Stramenopiles, Mor-
phology, Phylogeny.
Introduction
Newbya (formerly Achlya) is an important genus of the family Saprolegniaceae (Sapro-
legniales, Oomycota) that belongs to the monophyletic Kingdom Straminipila (Phylum
Heterokonta) (Spencer et al. 2002, Beakes et al. 2014, Steciow et al. 2014). Members of
© 2019 J. Cramer in Gebrüder Borntraeger Verlagsbuchhandlung, Stuttgart, www.borntraeger-cramer.de
Germany DOI: 10.1127/nova_hedwigia/2019/0537 0029-5035/2019/0537 $ 3.25
eschweizerbart_xxx
82 M.K. Dubey et al.
Newbya are characterized by Achlya-like typical sympodial type profuse branching, coe-
nocytic mycelium which gives rise to long, cylindrical, and usually terminal zoosporan-
gia towards the end with achlyoid discharge type but with predominantly multi oospored
oogonia characterized by centric or subcentric oospores (Spencer et al. 2002, Beakes et
al. 2014). Most of the species in this genus are adapted to living in aquatic habitats and
have specialized asexual propagules for effective dispersal in water called zoospores.
Apart from these morphological features, the genus mostly exhibits similar morphology
and lifestyles as the genus Achlya. Due to these similarities, for a long time, Newbya was
kept with Achlya under the family Saprolegniaceae (Leclerc et al. 2000). The Newbya
was erected from large paraphyletic genus Achlya based on molecular studies (Inaba &
Tokumasu 2002, Spencer et al. 2002), and Spencer et al. (2002) advocated for transferring
those species with centric or subcentric oospores from Achlya to a new genus, Newbya,
leaving remaining species having eccentric oospores in Achlya. In addition, the earlier
classification systems treated these fungal-like organisms having morphological similar-
ity with fungi as a basal fungal lineage but ultrastructural, biochemical and molecular
studies proposed that they are phylogenetically distinct to true fungi and are much closer
to chromophyte algae and other heterokont protists (Alexopoulos et al. 1996, Guarro et
al. 1999, Baldauf et al. 2000, Paul & Steciow 2008).The genus Newbya is globally dis-
tributed and currently represented by around 12 valid species (Spencer et al. 2002, Ste-
ciow et al. 2014). However, according to Khulbe (2001) the total number of reported
Newbya species from India is around eight. Members of the genus Newbya are mostly
saprobic on a wide variety of substrata serving simply as decomposers of organic materi-
als in all types of freshwater and soil ecosystems worldwide. However, some members
under favorable conditions are pathogens or necrotrophs as well, with diversified host
ranges affecting farmed and wildlife populations of aquatic animals (Dubey et al. 2018).
Despite these vital roles, the taxonomy and classification of the genus are still largely
based on a small number of morphological and ecological characters. The morphological
plasticity and ambiguous data regarding the proper identification of these zoosporic or-
ganisms, often results in error and confusion with other species of Saprolegnia and
Achlya. Several taxa are easily confused by their shared morphological features, and the
relationships between genera are inadequately recognized and in need of more critical
study. A morphological and molecular approach is required for a more accurate identifica-
tion of these pseudo-fungi. Nevertheless, the status of these stramenopiles in India needs
to be revised based on molecular analysis. Therefore, investigations on members of New-
bya have become of increasing interest, especially those focused on the identification and
characterization of species of these genera. Continued research on this economically im-
portant group of water-molds is therefore essential. India with all its available rivers,
lakes, and other fresh and saline waters sources seems an interesting place for the inves-
tigation of these fungal-like zoosporic organisms. Nevertheless, still not much is known
about their role, occurrence, and abundance of water-molds in this country.
As mentioned above, some Newbya species have received attention in India in the past,
however, still very little is known about diversity and occurrence of this group of water-
molds. To fill up this lacunae in the existing knowledge, in this paper, the authors report
a distinct and interesting Newbya species isolated from the yet unexplored dry tropical
eschweizerbart_xxx
First report of Newbya recurva (Saprolegniaceae) from India 83
forest of Chandra Prabha Wildlife Sanctuary, Chandauli district, Uttar Pradesh, India.
Despite its ecological and potential value, Chandra Prabha Wildlife Sanctuary has never
been explored for its mycobiota, particularly the water-molds. Through morphological
analysis, the identity of the specimen was determined to be Newbya recurva (Cornu) Dick
and Spencer. Thus, the objective of this study was to contribute to the knowledge of New-
bya species in India by giving particular emphasis to their taxonomical/morphological
characteristics having special features of N. recurva. The current study was undertaken to
evaluate this species based on morphological characters and phylogenetic analysis of the
ITS-rDNA region. Illustrated accounts are provided for the taxa, which are compared
with morphologically and phylogenetically related taxa.
Materials and methods
Sample collection and isolation
Water and soil samples with some organic debris were collected at random in separate
sterile labeled Ziploc bags from Rajdari waterfall, Chandra Prabha Wildlife Sanctuary,
Chandauli district, Uttar Pradesh, India (24°94'68" N and 83°16'86" E) during the end of
winter period (February 2015). The collected samples were brought to the laboratory,
processed by standard baiting technique using Spirulina flakes as baits in sterilized Petri
dishes following the methods outlined by Johnson (1956) and Sparrow (1960). The Petri
dishes containing water culture were incubated in the dark for a week at room tempera-
ture (20–25 °C). Within this incubation period, the baits were periodically examined un-
der the light microscope and when the growth of fungal-like zoosporic organisms was
observed on the baits, the small bits of colonized baits were washed thoroughly in sterile
distilled water and placed in another sterile Petri dishes containing sterile distilled water
and fresh sterile baits in order to obtain sister colonies. The current process was repeated
thrice and when mycelia growth became noticeable on the baits, the colonized baits were
re-washed several times in sterile distilled water and transferred onto PYG agar supple-
mented with penicillin G and streptomycin sulfate (300 ppm each) to obtain an axenic
culture that was maintained at 25 °C. After incubation and growth, the pure cultures were
established by dissecting out a block of agar (5mm in diameter) from the advancing edge
of the 4-day old colony and repeated reculturing on fresh PYG agar plates. Measurements
and observations were made on those colonies. The pure culture was transferred every 15
days and preserved for long term storage on sterile Spirulina flakes at 4–8 °C.
Identification using morphological characters
The identification of the isolate was performed on the basis of its morphological traits
using vegetative and reproductive characteristics on Spirulina flakes (baits), widely used
as a fish feed in aquaculture, under a light microscope. In brief, the mycological evalua-
eschweizerbart_xxx
84 M.K. Dubey et al.
tion and identification of the isolate was carried out on the basis of vegetative organs
(shape and size of the hyphae), asexual organs (shape and size of zoosporangium and
spores, their formation, patterns of discharge and germination) and sexual organs (pro-
duction, shape, structure and wall ornamentation of the oogonium, oospores per oogo-
nium, and type of antheridial branches) with the help of specialized monographs and de-
scriptions of Latham (1935), Coker & Matthews (1937), Johnson (1956), Dick (1959),
Seymour (1970), Dick (1973), Johnson & Seymour (1974), Johnson et al. (2002), Spen-
cer et al. (2002), and other relevant taxonomic literature containing original descriptions
of taxa. Sections were mounted in tapwater, in which all measurements were taken and
permanent mounts were prepared by fixing with formalin-acetic-alcohol and mounting in
lactophenol (Johnson 1956, Willoughby 1994). All morphological observations and
measurements were recorded, measured and photographed with a light microscope
(Dewinter microscope). The preserved specimen was deposited at Laboratory of Myco-
pathology and Microbial Technology, Centre of Advanced Study in Botany, Institute of
Science, Banaras Hindu University, Varanasi, India with the collection no. 147.
Genomic DNA extraction
For DNA extraction, the pure culture of the isolate was grown at pH 5.6 for 7 to 10 days
in Potato Dextrose Broth (PDB) at 25 ± 1 °C in a shaker incubator. The pellet containing
the mycelium was filtered through filter paper (Whatman no.1) and the collected total
genomic DNA was extracted using the Cetyltrimethyl Ammonium Bromide (CTAB)
method with minor modifications (Doyle & Doyle 1987). Approximately 0.5 g mycelium
mat was macerated in a pestle and mortar under liquid nitrogen, transferred to 10 ml of
DNA extraction buffer [0.1 M Tris (pH 8.0), 1.5 M NaCl, 0.01 M EDTA (pH 8.0)] and
kept at 65 °C for one hour with occasional stirring to break up the cells. Equal volumes of
chloroform/isoamyl alcohol (24:1) were added. The contents of the tube were vortexed
and mixed properly by inversion, followed by centrifugation. The upper aqueous phase
obtained by precipitation with the 0.6th volume of ice-cold isopropanol was recentrifuged.
The resulting precipitate was washed with 70% ethanol and air dried at room temperature.
Finally, the extracted DNA pellet was dissolved in 50 μl of 1X TE buffer and stored at
–20 °C for further use.
PCR amplification and sequencing of the ITS region
The identity of the isolate was confirmed by sequence analysis of rDNA internal tran-
scribed spacer (ITS) regions containing ITS1 and ITS2 and intervening 5.8 rDNA using
the primer pair ITS4 (5'TCCTCCGCTTATTGATATGC3')/ITS6 (5'GAAGGTGAA-
GTCGTAACAAGG3') (Cooke et al. 2000). The final volume of the PCR reaction
consisted of 50 μL reaction mixture, including 200 ng of each primer, 2 μl of each dNTP
(10 mM), 50 ng of DNA, 0.5 μl of Taq polymerase (3U/μl), and 0.5 μl of reaction buffer
eschweizerbart_xxx
First report of Newbya recurva (Saprolegniaceae) from India 85
(10X). The reaction mixtures were put through one cycle of the following in a DNA ther-
mal cycler: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation
at 94 °C for 30 sec, primer extension at 52 °C for 30 s, elongation at 72 °C for 45 s, with
a final 7 min extension step at 72 °C and a 10 °C hold.
The presence of successful amplification was checked on 1.2% (w/v) agarose gels by
electrophoresis, stained with ethidium bromide (5 mg/ml) in a TAE (1X) as the running
solution for 30 min. The electrophoretic migration was carried out during 2 hrs under an
80 V. The amplified products were visualized and photographed under UV light to check
success in PCR amplification. DNA sequencing was carried out at Chromous Biotech
Pvt. Ltd. (Bengaluru, India). In order to minimize sequencing errors, both strands were
sequenced using the chain termination method via an ABI 3500XL Genetic Analyzer
(Applied Biosystems Inc., Foster City CA, USA) and the sequences derived were finally
deposited in the GenBank database with accession no. MH685195.
Sequence alignment and phylogenetic analysis
The phylogenetic relationship of our isolate was established with species and isolates of
Achlya sensu lato based on sequence analysis of ITS-rDNA repeats. The BLASTn pro-
gramme was performed to evaluate the relationship of our isolate with other closely re-
lated organisms based on percent identity, maximum scores, query coverages and E-val-
ues at the public databases Genbank (http://www.ncbi.nem.nih.gov). The sequences hav-
ing good score matches with our target sequence were further aligned using Clustal W
and edited using BioEdit version 7.0.2 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html)
as implemented in MEGA 6.0. The best-fitting model for sequence evolution (GTR+gamma
distributed rate variation) was selected using the Akaike Information Criterion imple-
mented in jModeltest 2.1.10 (Darriba et al. 2012). A maximum likelihood phylogeny was
then estimated using PhyML 3.2 (Guindon et al. 2010) with 10 random searches to find
the best topology and 1,000 bootstrap pseudo-replicates to measure clade support.
Results
Description of the taxa
Newbya recurva (Cornu) M. W. Dick & Mark A. Spencer, Mycological Research 106 (5):
559. 2002. Fig. 1A–L
= Achlya recurva Cornu, Ann. Sci. Nat. Bot. (5 sér) 15: 22. 1872.
= Achlya papillosa Humphrey, Trans., Am. Philos. Soc., n. s., 17: 125. 1893.
= Achlya spiracaulis T. W. Johnson, Mycologia, 41: 678. 1949
eschweizerbart_xxx
86 M.K. Dubey et al.
Morphology
Newbya recurva is monoecious, saprobic and fast growing on Spirulina flakes (bait). A
three day-old colony on bait is about 1.2–1.5 cm in diameter with diffuse, moderately
extensively branched mycelium having dense hyphae near the Spirulina flakes. Principal
hyphae up to 25 μm thick at the base, slender and moderately branched. Gemmae rarely
present, often obovate or short cylindrical; usually single and intercalary or terminal.
Zoosporangia developed at the end of hyphae, usually terminal, abundant, clavate or fusi-
form, sometimes cylindrical, straight or curved, or with only the distal curve or irregular,
80–300 × 15–30 μm in diameter, tapering toward the end/apex, occasionally attenuated
into a long, narrow discharge tube; sympodial renewal or infrequently in basipetalous
succession; achlyoid zoospore discharge with compact clump formation (mass amor-
phous encysted in a cluster), persistent at the tip/top or exit pore of zoosporangia (from
both primary and secondary zoosporangia); in some cases compact clump not formed,
and zoosporangia releasing its amorphous protoplasm (aplanoid discharge). Encysted
zoospores or primary spore cysts, 10–12 μm diameter. Oogonia very abundant, spherical
or rarely sub-globose, extreme rarely dolioform or irregular; usually lateral, rarely termi-
nal, stalked, unpitted; wall provided with densely or sparsely externally ornamented, nu-
merous, elongated, stout, symmetrically arranged conic-truncate projections/protrusions
(the autapomorphic or hallmark feature), thin-walled at the apex but not bifurcated, cren-
ulate or papillate. Oogonia 50–90 μm in diameter, including the oogonial wall projections
(ornamentations). The projections themselves about 6–16 μm long and 6–15 μm wide at
the base. Oogonial stalks mostly short, stout, unbranched, or sometimes with one or two
papillate branches, straight, curved, or often recurved, frequently irregular, rarely twisted
or coiled, the length of stalks 0.75–1.5 times diameter of oogonium, a few stalks were
slightly longer. Antheridium with antheridial branches simple, short, rarely long, gener-
ally curved or bent, irregular, rarely branched, predominantly androgynous or rarely mo-
noclinous, extreme rarely diclinous; persisting. Antheridial cells simple, clavate or cylin-
drical, rarely irregular, often bent or curved, laterally or apically applied to the oogonium,
seldom in digitate fashion, tube non-persistent or persistent fertilization. Oospores not
always maturing, spherical, ellipsoidal or flattened adjacent to neighboring ones; centric
or subcentric, borne inside and usually 1–5 per oogonium, extreme rarely more than 5,
generally filling it; when single 30–40 μm in diameter, otherwise 25–35 μm diameter;
germination not observed; oospore wall about 2.5 μm thick (Fig. 1).
Culture characteristics: Colonies on PYG agar were fast growing, reaching 72–75 mm
in diameter after 4 days of incubation, with an average growth rate of 16.6 mm at 25–
28 °C. Colonies medium sparse, circular, puffy, whitish in color. Mycelium aseptate,
abundantly branched hyphae, partly immersed in the medium, partly superficial, with
tufting; not producing any kind of pigmentation in PYG agar.
Material examined: Chandra Prabha Wildlife Sanctuary (Chandauli district, Uttar
Pradesh, India), water and soil samples from Rajdari waterfall. February 2015, saprobic
on Spirulina flakes, No. 147 culture; GenBank accession no. MH685195.
Habitat: Fresh water streams rich in organic matter (leaves, branches, and litter).
eschweizerbart_xxx
First report of Newbya recurva (Saprolegniaceae) from India 87
Distribution: UK, Brazil, Argentina, Czechoslovakia, France, India, China, USA, Rus-
sia, Africa, Australia, Iceland, Denmark, Germany, Japan and Lithuania.
Notes: Within the members of Saprolegniaceae, this saprotrophic species is most easily
recognized by the presence of several striking feature such as oogonia external wall orna-
mented thin and truncate protrusions/decorations (hallmark feature), large subcentric
oospores and androgynous antheridia, which were observed in the isolated specimens,
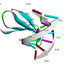
Fig. 1. Newbya recurva: A – Young vegetative thallus B – Aseptate and hyaline hyphae with with
primary and secondary sporangia renewed sympodially C–E – Maturing oogonia producing conic-
truncate projections F–G – An externally and heavily ornamented oogonium with conic-truncate
projections and androgynous antheridium H–J – A stalked oogonium with subcentric oospores and
androgynous antheridium K–L – A multisporus degenerated and discharged oogonium on a curved
stalk. Bars = 100 μm for A–D; 50 μm for E–L.
eschweizerbart_xxx
88 M.K. Dubey et al.
agreeing with the description of Johnson et al. (2002). The extensive perusal of literature
revealed that N. recurva was mostly recovered on hemp seed. However, it was never been
recovered using Spirulina flakes (over 1 mm diameter) as a bait. On this bait, the speci-
men grew abundantly producing the sex organs, particularly the oogonia in such an abun-
dance as to make the entire colony appear to be composed of ornamented oogonia within
4–5 days.
The comprehensive morphological description suggests that N. recurva is apparently
strikingly similar with A. primoachlya in the configuration of the oogonial wall. Johnson
et al. (2002) believed that the apex of most of the wall protrusions (ornamentations) in
both the taxa is thin and truncate. In addition, the origin of the antheridial branch in both
the taxa is also similar as is the predominant oospore number and its size. However, there
are certain contrasting known characters that seem to be sufficiently distinct from one
another to warrant their separation on several grounds. In general, the wall protuberances
(ornamentations) in N. recurva are more numerous and much more prominent than those
in A. primoachlya. On the other hand, the oogonia of the latter are more likely to be papil-
late than are those of the former. Further, in comparison to N. recurva, the strongly curved
oogonial stalks and thraustothecoid sporangia are never been reported in A. primoachlya.
Thus, our specimen agrees in all details with the above described characteristics but sepa-
rated/variable largely from their observations on the basis of oospore number (1–5) and
relatively smaller oogonia size.
Molecular characterization and phylogenetic studies
The molecular characterization based on ITS sequences analysis identified the queried
sequence as Newbya recurva (MH685195.1). The BLASTn annotation done for the que-
ried sequence revealed the first significant hit with maximum identity and percent se-
quence similarity with isolate A. recurva (HQ643107.1; maximum identity 99%; query
cover 96%; E-value 0.0). The phylogenetic analysis based on maximum likelihood re-
vealed a monophyletic N. recurva with three strains of A. recurva with high bootstrap
support (99%) (Fig. 2). This clade was sister to A. bisexualis, but with no substantial
bootstrap support. Overall, the phylogenetic analysis of species such as A. crenulata, A.
debaryana, and A. racemosa confirmed that Achlya is indeed polyphyletic, which sup-
ports the prevailing idea (Spencer et al. 2002) that A. recurva and N. recurva should be
considered a single species in the genus Newbya.
Discussion
In the framework of a project of zoosporic fungi and fungal-like organism exploration
through Chandra Prabha Wildlife Sanctuary, a large number of specimens were col-
lected. Through morphological examination of our collections, we came across a surpris-
ingly common species of the genus Newbya. We isolated, described, illustrated and pro-
eschweizerbart_xxx
First report of Newbya recurva (Saprolegniaceae) from India 89
vided a key with a synopsis to this species to facilitate its identification. This species is so
far reported by Saksena and Rajagopalan (1958), Manoharachary (1979), Chowdhry &
Agarwal (1980), Manoharachary & Rao (1981), and Bhairabnath & Manoharachary
(1985) from various locations in India but those reports were not supported by molecular
analysis, so these reports should be considered tentative due to the lack of molecular evi-
dence. Further, these prior works suffer from many inconsistencies including observation
on reproductive apparatus (asexual and sexual organs) and their illustrations, due to
which their authenticity is doubtful. The results of our specimen’s morphological charac-
Fig. 2. Maximum likelihood tree inferred from ITS-rDNA sequences of Newbya. Sequence obtained
in this study is underlined. The maximum likelihood tree was inferred using PhyML with a GTR +
gamma rate variation model. Shown above nodes are proportion of bootstrap replicates supporting
each node; only values above 50% are shown.
eschweizerbart_xxx
90 M.K. Dubey et al.
terization coincided with the description of Newbya recurva (syn. Achlya recurva) re-
ported by Latham (1935), Coker & Matthews (1937), Steciow (1997), Johnson et al.
(2002), and Spencer et al. (2002). Currently, only three ITS sequences of A. recurva are
available in GenBank; our sequence (MH685195) is the fourth. Two of the three se-
quences are of unknown geographic provenance, and the third is from Iran. Thus the
current report is the first report of Newbya recurva based on morphological and molecular
analysis of ITS-rDNA in India.
In India, since the previous contribution of Khulbe (2001), the water-molds have been
poorly studied and neglected. In addition, basic knowledge of their distribution and eco-
logical roles is still in its infancy. Although genera of particular groups of Oomycota
(Peronosporales) are well known and frequently investigated organisms, many other gen-
era (especially in lesser known orders) have remained obscure. The extensive perusal of
literature revealed that more than twelve Newbya species have been reported globally.
Newbya spp. are common inhabitants of aquatic environments; however, there are few
quantitative studies made for Indian streams. In this sense, nearly eight described, ac-
cepted species of Newbya, namely, N. pascuicola (Basionym: Aplanopsis spinosa), N.
apiculata (Basionym: Achlya apiculata), N. megasperma (Basionym: Achlya mega-
sperma), N. oblongata (Basionym: Achlya oblongata), N. oligacantha (Basionym: Achlya
oligacantha), N. polyandra (Basionym: Achlya polyandra), N. recurva (Basionym:
Achlya recurva; Syn.: Achlya papillosa) and N. stellata (Basionym: Achlya stellata) are
known and reported from India (Khulbe 2001). However, none of the above mentioned
species have been molecularly identified. This lacunae in the current data clearly high-
lights the need for further study. Nevertheless, there are still numerous species of Newbya
that unknown to science in India. It implies that there is a potential for describing several
new species or new records of Oomycota (Saprolegniales) from largely unexplored areas
in India. Furthermore, most studies dealing with water-molds have provided extensive
inventories of taxa from specific sites or geographic regions, but often without character-
izing the microhabitat, determining frequencies of occurrence or relative abundance of
species (Dubey et al. 2018). Still very little is known regarding the role that most Newbya
play in native communities in other parts of the world. Despite the economic and eco-
logical importance of these stramenopiles, much remains to be learned about their bio-
logy, particularly their host and geographic ranges. Hence, we felt it was imperative to
study the morphological features of N. recurva. The classification of Newbya has been a
major challenge due to the lack of a clear understanding of the important morphological
characters, extensive morphological overlap among recognized species as well as a lack
of reference strains. Classification in the genus Newbya has been largely based on mor-
phology, with oogonial characters being considered as important in distinguishing species
and closely related genera. However, inadequate attention to water-mold systematics has
led to confusion in their taxonomic, and numerous taxa might have been characterized
incorrectly either in terms of sexual or of asexual features. As a partial remedy to what
appears to be a persistent problem/mammoth task in the systematics of water-molds,
Johnson et al. (2002) mentioned certain criteria that should be taken in consideration be-
fore any water-mold is assigned to the genus. However, nowadays resolution of taxonomy
using nucleotide sequences of the internal transcribed spacer (ITS) of the rDNA region is
eschweizerbart_xxx
First report of Newbya recurva (Saprolegniaceae) from India 91
considered the best tool for rapid and accurate identification of water-mold isolates (Ste-
ciow et al. 2014). The present note is the fourth attempt by the author on more poorly
understood representatives water-mold of India (Dubey & Upadhyay 2013, Dubey et al.
2016, 2018) to provide at least a modicum of their taxonomic study, and the first report
by morpho-molecular analysis of Newbya in India.
Conclusion
An isolate of N. recurva was identified using morphology as the key character for taxo-
nomic circumscription and DNA sequencing of the internal transcribed spacer (ITS) re-
gion of rDNA. Short descriptions, illustrations, and notes on species morphological vari-
ability for their identification using solely morphological and reproductive characters on
Spirulina flakes bait were discussed. The results of the present study provide an exhaus-
tive morpho-taxonomical characterization, distinguishing features and ecological/distri-
butional data of N. recurva, a critical species of fungi-like zoosporic organism in fresh-
water bodies of India. The current communication could be likely considered as the first
scientific report supported with molecular analysis pertaining to the occurrence/presence
of N. recurva in India, which is also the first record of it on Spirulina flakes used as a bait.
Undoubtedly, there are many additional Newbya species that are yet to be reported. Alter-
native culturing techniques, growth media, and growth conditions in future investigations
will reveal additional Newbya species and ultimately add to our understanding of the
water-molds associated with different water bodies in India.
Acknowledgments
We are thankful to the Head, Department of Botany and Programme Coordinator, Centre
of Advanced Study in Botany, Banaras Hindu University for providing necessary facili-
ties and support. The author Manish Kumar Dubey wishes to thank the Centre of Ad-
vanced Study in Botany, Banaras Hindu University, India for providing financial assis-
tance in form of a Senior Research Fellowship (SRF) during the course of study.
References
Alexopoulos, C.J., Mims, C.W. & Blackwell, M. (1996): Introductory mycology. 4thed. – John
Wiley, New York.
Baldauf, S.L., Roger, A.J., Wenk-Siefert, I. & Doolittle, W.F. (2000): A kingdom-level phylogeny
of eukaryotes based on combined protein data. – Science 290: 972–977.
Beakes, G.W., Honda, D. & Thines, M. (2014): Systematics of the Straminipila: Labyrinthulomy-
cota, Hyphochytriomycota and Oomycota. – In: McLaughlin, D.J. & Spatafora, J.W. (eds.): The
Mycota VIII Part A; p. 39–97. Springer-Verlag, Berlin.
eschweizerbart_xxx
92 M.K. Dubey et al.
Bhairabnath, D. & Manoharachary, C. (1985): Mycoecological studies of two water bodies from
Andhra Pradesh, India. – Indian J. Bot. 8: 69–78.
Chowdhry, P.N. & Agrawal, G.P. (1980): Studies on distribution of some aquatic fungi in India. –
Indian Phytopathol. 33: 107–109.
Coker, W.C. & Matthews, V.D. (1937): Blastocladiales, Monoblepharidales, Saprolegniales. –
North Am. Flora 2: 15–75.
Cooke, D.E.L., Drenth, A., Duncan, J.M., Wagels, G. & Brasier, C.M. (2000): A molecular phylo-
geny of Phytophthora and related Oomycetes. – Fungal Genet. Biol. 30: 17–32.
Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012): jModelTest 2: more models, new heu-
ristics and parallel computing. – Nat. Methods 9(8): 772.
Dick, M.W. (1959): The occurrence and distribution of Saprolegniaceae in certain soils of south
east England, with taxonomic notes of little known species. – PhD thesis, Queen Mary College,
University of London.
Dick, M.W. (1973): Saprolegniales. – In: Ainsworth, G.D., Sparrow, F.K. & Sussman, A.S. (eds.):
The Fungi: An Advanced Treatise; p. 113–144. Academic Press, New York.
Doyle, J.J. & Doyle, J.L. (1987): A rapid DNA isolation procedure for small quantities of fresh leaf
tissue. – Phytochem. Bull. 19: 11–15.
Dubey, M.K. & Upadhyay, R.S. (2013): Isolation and characterisation of some Indian Hyphochy-
triomycetes. – Int. Res. J. Biol. Sci. 2: 31–34.
Dubey, M.K., Zehra, A., Meena, M. & Upadhyay, R.S. (2016): Taxonomic notes on Allomyces neo-
moniliformis (Blastocladiaceae) isolated from Nainital lake, Uttarakhand, India. – Vegetos 29:
1–7.
Dubey, M.K., Zehra, A., Meena, M. & Upadhyay, R.S. (2018): Taxonomic note on a rare fish infect-
ing freshwater mould Achlya ambisexualis Raper 1939 (Achlyaceae) isolated from Chan-
draprabha dam, Uttar Pradesh, India. – Indian J. Fish. 65: 71–78.
Guarro, J., Gene, J. & Stchigel, A. (1999): Developments in fungal taxonomy. – Clinical Microbiol.
Rev. 12: 454–500.
Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W., et al. (2010): New algorithms
and methods to estimate maximum-likelihood phylogenies: Assessing the performance of
PhyML 3.0. – Syst. Biol. 59(3): 307–321.
Inaba, S. & Tokumasu, S. (2002): Phylogenetic relationships between the genus Saprolegnia and
related genera inferred from ITS sequences. – Abstracts of the 7th international mycological
congress, Oslo IMC7, p. 687.
Johnson, T.W. (1956): The genus Achlya: morphology and taxonomy. – University of Michigan
Press, Ann Arbor.
Johnson, T.W. & Seymour, R.L. (1974): Aquatic fungi of Iceland: comparative morphology of
Achlya spiracaulis and Achlya papillosa. – Nova Hedwigia 25: 433–449.
Johnson, T.W., Seymour, R.L. & Padgett, D.E. (2002): Biology and systematics of Saprolegniaceae.
– University of North Carolina, Wilmington.
Khulbe, R.D. (2001): A manual of aquatic fungi. – Daya Publisher House, New Delhi, India.
Latham, D.H. (1935): Achlya recurva Cornu from North Carolina. – J. Elisha Mitchell Sci. Soc. 51:
183–187.
Leclerc, M.C., Guillot, J. & Deville, M. (2000): Taxonomic and phylogenetic analysis of Saproleg-
niaceae (Oomycetes) inferred from LSU rDNA and ITS sequence comparisons. – Antonie Van
Leeuwenhoek 77: 369–377.
Manoharachary, C. (1979): Taxo-ecological studies on some freshwater pond mud fungi of Hy-
derabad, India. – Nova Hedwigia 63: 197–207.
Manoharachary, C. & Rao, R. (1981): Seasonal variation and distribution of fungi in two freshwater
ponds of Andhra Pradesh, India. – Proc. Indian Acad. Sci. 90: 237–243.
Paul, B. & Steciow, M.M. (2008): Achlya spiralis, a new aquatic oomycete with bent oogonial
stalks, isolated from the Burgundian region of France. – FEMS Microbiol. Lett. 284: 120–125.
eschweizerbart_xxx
First report of Newbya recurva (Saprolegniaceae) from India 93
Saksena, S.B. & Rajagopalan, C. (1958): Studies on aquatic fungi of Saugar. – J. Univ. Saugar IIB
7: 7–20.
Seymour, R. (1970): The genus Saprolegnia. – Nova Hedwigia 19: 1–124.
Sparrow, F.K. Jr. (1960): Aquatic Phycomycetes. – University of Michigan Press, Ann Arbor, Mi-
chigan.
Spencer, M.A., Vick, M.C. & Dick, M.W. (2002): Revision of Aplanopsis, Pythiopsis, and ‘subcen-
tric’ Achlya species (Saprolegniaceae) using 18S rDNA and morphological data. – Mycol. Res.
106: 549–560.
Steciow, M.M., Lara, H., Paul, C., Pillonel, A. & Belbahri, L. (2014): Multiple barcode assessment
within the Saprolegnia-Achlya clade (Saprolegniales, Oomycota, Straminipila) brings order in
a neglected group of pathogens. – IMA Fungus 5: 439–448.
Steciow, M.M. (1997): The occurrence of Achlya recurva (Saprolegniales, Oomycetes), in hydro-
carbon polluted soil from Argentina. – Rev. Iberoam. Micol. 14: 135–137.
Willoughby, L.G. (1994): Fungi and fish diseases. Stirling, Pisces Press Publication. – Kasetsart J.
(Nat. Sci.) 46: 91–97.
Manuscript received: December 13, 2018
Accepted: February 14, 2019
Responsible editor: J. Błaszkowski
eschweizerbart_xxx