Content uploaded by Merje Toome-Heller
Author content
All content in this area was uploaded by Merje Toome-Heller
Content may be subject to copyright.
Meredithblackwellia eburnea
gen. et sp. nov.,
Kriegeriaceae fam. nov. and Kriegeriales ord. nov.—toward
resolving higher-level classification in Microbotryomycetes
Merje Toome
1,2
Department of Plant Pathology and Crop Physiology,
Louisiana State University Agricultural Center,
Baton Rouge, Louisiana 70803
Robert W. Roberson
School of Life Sciences, Arizona State University, Tempe,
Arizona 85287
M. Catherine Aime
1
Department of Plant Pathology and Crop Physiology,
Louisiana State University Agricultural Center,
Baton Rouge, Louisiana 70803
Abstract
:A field survey of ballistosporic yeasts in a
Neotropical forest yielded a new species isolated from
a fern leaf. The isolate is a cream-colored butyrous
yeast that reproduces by budding. Budding occurs at
both the apical and basal cell poles; occasionally
multiple budding events co-occur, giving rise to
rosette-like clusters of cells at both poles of the yeast
mother cell. DNA sequences of large and small
subunit and the internal transcribed spacer regions
of the nuclear ribosomal DNA cistron indicated an
affinity to Microbotryomycetes, Pucciniomycotina. A
new genus,
Meredithblackwellia,
is proposed to accom-
modate the new species,
M. eburnea
(type strain
MCA4105). Based on phylogenetic analyses,
Meredith-
blackwellia
is related to
Kriegeria eriophori
, a sedge
parasite, to an aquatic fungus
Camptobasidium hydro-
philum
and to several recently described anamorphic
yeasts that have been isolated from plant material or
psychrophilic environments. Morphological and ul-
trastructural studies confirm the relatedness of
M.
eburnea
to these taxa and prompted the re-evaluation
of higher-level classification within Microbotryomy-
cetes. We propose here a new order, Kriegeriales, and
place two families, Kriegeriaceae fam. nov. and
Camptobasidiaceae R.T. Moore, within it. Our study
re-emphasizes the need for systematic revision of
species described in
Rhodotorula
.
Key words:
basidiomycete yeasts, fungal taxono-
my, phylloplane, simple septate basidiomycetes
INTRODUCTION
The external surface of aerial plant parts (phyllo-
sphere or phylloplane) accommodates a highly
diverse microbial community. A high number of
various prokaryotic and eukaryotic organisms have
been found to colonize plant surfaces, showing
adaption to this habitat to be a successful evolutionary
trend that has taken place frequently and indepen-
dently several times (Andrews and Harris 2000).
Among fungi, yeasts are the most commonly isolated
active phylloplane organisms; filamentous fungal
species, on the other hand, are recovered more often
from plant surfaces in the dormant spore stage
(Nakase 2000, Whipps et al. 2008). It has been shown
that many phylloplane yeasts could protect plants
from pathogens and therefore promote plant growth
(e.g. McCormack et al. 1994, Buck 2002); neverthe-
less, some of them are thought to be parasitic.
Because only a few species have been studied in
detail, the ecology of the majority of yeasts on plants is
poorly understood and their composition may signif-
icantly vary depending on climate conditions or the
surrounding microbial communities (Andrews and
Harris 2000).
Several studies examining the diversity of yeasts on
plants have found that the most frequently recovered
species are yeasts from Pucciniomycotina, mostly
belonging to the genera
Sporobolomyces
,
Rhodotorula
and
Bensingtonia
(e.g. Nakase 2000). Most of these
have ballistosporic spore discharge, a feature charac-
teristic of many basidiomycetes (Kirk et al. 2008),
including yeasts and species with yeast states in
Pucciniomycotina. Historically these yeasts have been
placed into form-genera based on their carbon
assimilation abilities and colony pigmentation (Kurtz-
man et al. 2011). Recent advances in reconstructing
the phylogenetic relationships of Pucciniomycotina
(e.g. Aime et al. 2006) now allow the application of
molecular phylogenetics to facilitate the integration
of anamorphic yeasts within the teleomorph-based
classification. For example, species placed in
Spor-
obolomyces
occur across most of the yeast-forming
Pucciniomycotina classes and species placed in
Rhodotorula
can be found in Ustilaginomycotina as
well as Pucciniomycotina (Sampaio 2004, Scorzetti et
al. 2002). The type species for both
Rhodotorula
(
R.
glutinis
(Fresen.) F.C. Harrison) and
Sporobolomyces
Submitted 11 Jul 2012; accepted for publication 21 Sep 2012.
1
Current address: Department of Botany and Plant Pathology,
Purdue University, West Lafayette, Indiana 47907.
2
Corresponding author. E-mail: mtoome@purdue.edu
Mycologia,
105(2), 2013, pp. 486–495. DOI: 10.3852/12-251
#2013 by The Mycological Society of America, Lawrence, KS 66044-8897
486
(
S. salmonicolor
(B. Fisch. & Brebeck) Kluyver & C.B.
Niel) are now known to belong to Sporidiobolales in
Microbotryomycetes (Scorzetti et al. 2002), highlight-
ing the necessity for additional taxonomic studies that
will integrate the ex-
Sporobolomyces
s.s. and ex-
Rhodotorula
s.s. species into a natural classification.
Microbotryomycetes is the second largest class in
Pucciniomycotina with more than 200 described
species. Microbotryomycetes primarily contains the
‘‘anther smuts’’, smut-like species formerly classified
in Ustilaginomycotina, and numerous anamorphic
yeasts (Bauer et al. 2006). At present, the class
contains four orders: Microbotryales—predominantly
teliospore-forming plant parasites; Sporidiobolales
and Leucosporidiales—anamorphic or teliospore-
forming yeasts isolated from various habitats and
surfaces; and Heterogastridiales—hyphal fungi isolat-
ed from decaying plant material and mushrooms
(Aime et al. 2006, 2012). Although there have been
great improvements in circumscribing a natural
Microbotryomycetes over the past decade, the place-
ment of almost a quarter of the species within it
remains unresolved. For instance,
Kriegeria
,
Campto-
basidium
and
Colacogloea,
all monotypic genera, are
shown to be members of Microbotryomycetes (Aime
et al. 2006) but are still classified incertae sedis within
the class as are many anamorphic species, especially
those currently placed in either
Rhodotorula
or
Leucosporidium
. In this paper we describe a new yeast
genus and species that was isolated from a fern
phylloplane in western Guyana and provide a three-
locus phylogenetic analysis of Microbotryomycetes. As
a result, one new order and one new family,
Kriegeriales and Kriegeriaceae, are described, and
the higher-level placement of many species previously
placed incertae sedis is resolved.
MATERIALS AND METHODS
Sample collection and isolation.—
The yeast strain described
in this study (collection No. MCA4105) was isolated from
the leaf surface of an unidentified fern at a permanent base
camp in the Pakaraima Mountains in western Guyana
(5u18904.80N, 59u54940.40W; 710 m) on 28 May 2010. The
leaf was cut into small pieces that were attached with a thin
layer of petroleum jelly to the inner lid of a Petri dish
containing potato dextrose agar media (PDA) with added
chloramphenicol (1 mL L
21
), to avoid bacterial growth.
Plates were monitored daily by eye for presence of colonies,
which were transferred with sterile toothpicks into 2 mL
microtubes containing the same media. These isolates were
kept in the microtubes until transferred to laboratory
conditions, where pure cultures were streaked out and
stored on PDA at 4 C and as glycerol stocks at 280 C for
long-term storage. The culture is deposited in Centraalbur-
eau voor Schimmelcultures (CBS) Fungal Biodiversity
Centre, the American Type Culture Collection (ATCC)
and the Agricultural Research Service Culture Collection
(NRRL) under deposit Nos. CBS12589, ATCC MYA-4884
and NRRL Y-48821 respectively.
Physiological and morphological characterization.—
Physio-
logical, biochemical and morphological properties of the
isolate were determined following Kurtzman et al. (2011).
The fermentation of glucose and the assimilation of carbon
and nitrogen compounds were determined at 25 C over
4 wk. The effect of temperature was determined during 2 wk
at 4, 20, 25 and 30 C on agar plates containing yeast malt
agar (YMA) and cornmeal Agar (CMA) media. Morpholog-
ical characters of the isolate were observed on YMA and
CMA plates at 1, 2 and 3 wk after inoculations.
To describe microscopic characters, the culture was
incubated in YM broth and PDA media for 5 d and actively
budding yeast cells were viewed with a Zeiss (Carl Zeiss, Inc.,
Thornwood, New York) dissecting light microscope with a
403objective. For standard differential interference con-
trast (DIC) observations, cells from a 5 d old culture on PDA
were directly observed. For nuclear division observations,
actively growing cells from the same culture were fixed with
freshly prepared 4%formaldehyde in phosphate buffered
saline(PBS,0.05M,pH6.8)for30minatroom
temperature, washed in PBS, and stained in 49,6-diami-
dino-2-phenylindole (DAPI; Sigma, St Louis, Missouri) at
0.1 mg/mL in H
2
0 for 5 min. Cells were rinsed in H
2
0 and
mounted on glass slides in 90%glycerol—10%0.1 M PBS
(pH 8.6)—2%N-propyl gallate (Sigma) and viewed with
DIC and epifluorescence optics. All images were viewed on
an Axioscope microscope (Carl Zeiss, Inc.) and observed
with a Plan-Neofluor 1003/1.3 (oil immersion) objective
and captured with a Roper Cool SNAP ES digital camera
(Roper Scientific Inc., Tucson, Arizona) with MetaMorph
6.0/6.1 software (Universal Imaging Corp., Downingtown,
Pennsylvania).
To describe the ultrastructure of MCA4105, cells were
grown as monolayers on a thin, sterile, deionized dialysis
membrane overlying PDA at 23 C. After approximately 24 h
growth, cells and supporting membranes were trimmed with
a razorblade to approximately 5 35 mm, removed from the
agar surface and immediately cryofixed by plunging rapidly
into liquid propane cooled to 2186 C with liquid nitrogen
(Hoch 1986, Roberson and Fuller 1988). Cryofixed cells
were freeze substituted in acetone containing 2%OsO
4
at
285 C for 48–72 hr. While in the substitution solution, the
cells were slowly warmed to room temperature, rinsed with
100%acetone and infiltrated with epoxy resin. Infiltrated
cells were flat-embedded between a Teflon-coated glass slide
and Teflon strips and polymerized at 60 C for 24 h. After
resin polymerization, selected hyphae were sectioned on a
Leica Ultracut microtome (Leica Microsystems Inc., Ban-
nockburn, Illinois) and post stained for 5 min in 2%uranyl
acetate in 50%ethanol and for 3 min in lead citrate.
Sections were examined with a FEI CM12S transmission
electron microscope (TEM) (FEI Electronics Instruments
Co., Mahwah, New Jersey) at 80 kV coupled to a Gatan 689
CCD digital camera (1024 31024 pixel area; Gatan Inc.,
Pleasanton, California). For all imaging methods, final
TOOME ET AL.: HIGHER-LEVEL CLASSIFICATION IN MICROBOTRYOMYCETES 487
figures were constructed with Adobe Photoshop 7.0 (Adobe
Systems Inc., San Jose, California).
DNA extraction, sequencing and phylogenetic analysis.—
DNA
was extracted from colonies actively growing on PDA with
Promega Wizard Genomic DNA Purification Kit (Promega,
Madison, Wisconsin). PCR reactions were carried out in
25 mL reactions that contained 12.5 mL GoTaq Master Mix
(Promega), 1.25 mL each primer (10 mM), 9 mL molecular
grade water and 1 mL DNA template. Amplifications of the
internal transcribed spacer (ITS) region, large (LSU) and
small (SSU) subunit of the nuclear ribosomal DNA (rDNA)
were conducted with primer pairs ITS1F (Gardes and Bruns
1993)/ITS4 (White et al. 1990), 5.8SR/LR6 (Vilgalys and
Hester 1990) and NS1/NS4 and NS3/NS8 (White et al.
1990) respectively. Amplification conditions for the ITS
region were 5 min at 95 C followed by 35 cycles of 30 s at
94 C, 45 s at 45 C and 45 s at 72 C, ended with a 7 min
extension at 72 C. The same PCR conditions were used for
LSU and SSU, except that for both the elongation step was
extended to 1 min and for SSU annealing was conducted at
55 C for 30 s. Sequencing of amplified fragments was
performed by Beckman Coulter, Inc. (Danvers, Massachu-
setts) with the same primers that were used for amplifica-
tion. The SSU, LSU and ITS sequences of MCA4105 are
deposited in GenBank under accession numbers JX508797,
JX508798 and JX508799 respectively.
Sequences were edited with Sequencher 4.10.1 (Gene
Codes Corp.) and blasted in GenBank (www.ncbi.nlm.nih.
gov) with BLASTn for initial identification, which indicated
that the isolate belonged to Pucciniomycotina. Thereafter
the LSU sequence was analyzed within the dataset of Aime
et al. (2006), which supported placement within Microbo-
tryomycetes. New datasets were constructed for each
sequenced locus by aligning the closest available blast
matches with at least 94%identity and a selection of other
members of Microbotryomycetes. To determine the phylo-
genetic position within the Microbotryomycetes, the first
dataset combined all three loci for 16 Microbotryomycetes
species, including exemplars from each described order
and
Mixia osmundae
as an outgroup, because this is one
potential sister group to Microbotryomycetes in prior
studies (Aime et al. 2006). A second dataset was created
for family positioning containing LSU and ITS sequences of
18 taxa found to be the most closely related to MCA4105 by
BLASTn and prior phylogenetic analyses. SSU was not
included in this dataset due to lack of available sequences
for selected taxa.
Leucosporidium scottii
was included to test
monophyly of Kriegeriales, and
Microbotryum violaceum
was
chosen as an outgroup based on the results from analysis of
dataset 1. Sequences were aligned with the MUSCLE
algorithm in MEGA 5.0 (Kumar et al. 2008). In dataset 1,
the sequences of LSU, ITS and SSU were aligned along 587,
533 and 1349 base pairs respectively. For dataset 2,
ambiguously aligned regions were excluded from analyses
and therefore LSU and ITS sequences were aligned along
453 and 404 base pairs respectively. The alignments are
available in TreeBASE (treebase.org) under submission
number 13340. Details of sequences used for these analyses
are presented (SUPPLEMENTARY TABLE I).
The maximum likelihood (ML) analyses were conducted
in RAxML-HPC2 7.2.8 via the CIPRES Science Gateway
(Miller et al. 2010) using the 2k option for bootstrap
analysis. Bayesian posterior probability analyses were con-
ducted in MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003)
with parameters set to 5 000 000 generations, four runs and
four chains. The chains were heated to 0.25 and a stop value
of 0.01 was used. Maximum parsimony (MP) analyses were
conducted in PAUP* 4.0b10 (Swofford 2002) where the
support of the branching topologies derived from 1000
replicates with 10 random additions.
RESULTS
The isolated yeast is fast-growing, forming white
butyrous colonies on agar media (FIG. 1a). Micro-
scopic studies showed that MCA4105 reproduces by
budding, which occurs on a narrow base from each
pole, although occasionally lateral budding was
observed (FIG. 1b, h). Cells produce up to three buds
at the same time either on the same or opposite apex
and multiple budding events give rise to rosette-like
clusters (FIG. 1c–e). Ultrastructural and nuclear divi-
sion studies showed that during budding the nucleus
moves into the forming bud (FIG. 1f–i), and during
mitosis one spindle pole remains in the bud while the
other one migrates back into the mother cell (FIG. 1j,
k). Microscopy studies also showed that MCA4105
cells contain numerous vacuoles and lipid bodies
(FIGS. 1b, c, 2a). Additionally, ultrastructure studies
revealed subgloboid spindle pole bodies with an
internal flattened layer and numerous mitochondria
(FIG. 2a–c). In actively budding cells, increased Golgi
complexes and numerous vesicles were found in the
immediate proximity of the tip of the bud, and newly
forming buds were surrounded with a thick layer of
extracellular matrix (FIG. 2e, f). Budding is entero-
blastic, and after the bud matures a new membrane
and cell wall form to close the budding site (FIG. 2g–i).
The LSU sequence of MCA4105 shares 97%
identity (1035 of 1066 bp) with
Rhodotorula
sp.
FK.2.1. (GenBank No. FN400943; unpubl), isolated
from the nectar of
Viola
sp.inGermany,and
Rhodotorula
sp. KBP 3844 (No. FN400942; unpubl),
isolated from a lichen on
Picea abies
in Russia. The
SSU region shared highest identity (99%; 1558 of
1571 bp) with the plant parasite
Kriegeria eriophori
(DQ419918; Kumar et al. 2007) and 98%identity
(1542 of 1571 bp) with
Glaciozyma antarctica
(DQ785788; Matheny et al. 2006) and
Leucosporidium
sp. AY30 (GQ336996; unpubl), the latter two isolated
from ice and water. The closest identities in the ITS
region were with uncultured
Rhodotorula
isolates
(GU931760, GU931719; Amend et al. 2010) from
house dust in Canada that shared 84%identity (545
and 541 of 652 bp respectively), and
Rhodotorula
sp.
488 MYCOLOGIA
USM-PSY62 (HM545717; unpubl) from Antarctic sea
with 83%identity (540 of 652 bp). Phylogenetic
analyses place MCA4105 with
K. eriophori
in a lineage
that is distinct from the other described orders of
Microbotryomycetes (FIG. 3A) and sister to Campto-
basidiaceae (FIG. 3B). MCA4105 forms a distinct
lineage, sister to
Rhodotorula rosulata
(FIG. 3B) and
a new genus and species are erected to accommodate
this isolate.
TAXONOMY
Kriegeriales Toome & Aime, ord. nov.
MycoBank MB801220
Etymology:
The name of the order is derived from the
genus
Kriegeria
Bres. (Bresadola 1891), which contains the
earliest described known member.
Diagnosis:
Members of Pucciniomycotina, Micro-
botryomycetes, with simple pore septa and subgloboid
spindle pole bodies. Hyphae, when present, may have
clamp connections. Asexual reproduction occurs
mostly by polar budding or, in one species, by
production of aquatically adapted tetraradiate spores.
Teleomorph variable where known either aquatic or
plant parasitic and sorus-forming. Basidiospores are
sessile or singly formed on sterigmata, on transversely
septate basidia.
Known members of the Kriegeriales are plant
parasites, aquatic fungi or saprobes from cold,
temperate and tropical environments. The majority
of members are known as anamorphic non-pigment-
ed yeasts; a teleomorphic state is known only for
K.
eriophori
and
C. hydrophilum
. The order accommo-
dates two families: Kriegeriaceae Toome & Aime and
Camptobasidiaceae R.T. Moore.
Kriegeriaceae Toome & Aime, fam. nov.
MycoBank MB801221
Type: Kriegeria
Bres. (Bresadola 1891).
Etymology:
The name of the family is derived from the
genus
Kriegeria
.
Diagnosis:
Members of Pucciniomycotina, Micro-
botryomycetes, Kriegeriales, with simple pore septa
and subgloboid spindle pole bodies. Anamorphic
states grow as colorless to cream-colored yeasts that
reproduce most commonly by enteroblastic polar
budding and form chains or rosettes. The nuclear
division takes place in the daughter cell. Some
anamorphs form hyphae and pseudohyphae. Tele-
omorphic state, where known, hyphal, plant parasitic
and sorus-forming. Basidiospores form singly on
transversely septate metabasidium, which also may
produce yeast cells.
To date three genera are recognized:
Kriegeria
Bres.
and its anamorph,
Zymoxenogloea
D.J. McLaughlin
and Double´s, and
Meredithblackwellia
Toome & Aime.
Known species are
K. eriophori
(5
Z. eriophori
),
Rhodotorula glacialis
,
R. himalayensis
,
R. pilati
,
R.
psychrophenolica
,
R. psychrophila
,
R. rosulata
,
Rhodo-
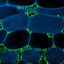
FIG. 1. Morphological characteristics of
Meredithblackwellia eburnea
(HOLOTYPE, MCA4105). a. 7 d old culture on YMA
media, showing the smooth butyrous culture with entire margins and single colonies with elevated center. Bar 51 cm. b.
Apically budding yeast cells and non-budding cells with vacuoles (asterisk) and lipid bodies (white arrow). Bar 510 mm. c.
Numerous cells forming a rosette. Vacuoles are noted (asterisks). Bar 510 mm. d. Apical budding (arrowheads) occurring
simultaneously from both poles. Bar 55mm. e. Three buds forming from the same site (arrowhead). Bar 55mm. f–k. Nuclear
division observed in the cells using epifluorescence microscopy and associated DIC images. Note the nucleus moving into the
bud (h, i) and then back to the mother cell (j, k). Bar 510 mm.
TOOME ET AL.: HIGHER-LEVEL CLASSIFICATION IN MICROBOTRYOMYCETES 489
torula
sp. KRP3844,
Rhodotorula
sp. FK.2.1,
Rhodotor-
ula
sp. CBS11784 and
Meredithblackwellia eburnea
.
Meredithblackwellia Toome & Aime, gen. nov.
MycoBank MB801222
Type: Meredithblackwellia eburnea
Toome & Aime, this
paper.
Etymology: Meredithblackwellia,
in honor of Meredith
Blackwell, Boyd Professor, Louisiana State University, for
her vast contributions to yeast research.
Diagnosis:
Members of Pucciniomycotina, Micro-
botryomycetes, Kriegeriales, Kriegeriaceae with sub-
globoid spindle pole bodies with a flat internalized
layer. Anamorphic yeasts that grow as cream-colored
butyrous colonies on PDA and YMA. Cells are
elongated, bud mostly on a narrow base from each
pole, and form rosettes of semi-attached cells. Cells
contain numerous vacuoles and lipid bodies and
nuclear division occurs inside the bud. Teleomorph
not known.
FIG. 2. Ultrastructural characters of
Meredithblackwellia eburnea
(HOLOTYPE, MCA4105). a. Mature yeast cell with several
vacuoles (V) and lipid bodies (arrows). Bar 51mm. b. Nucleus (N) and nucleolus (NU) and a mitochondrion (M). Bar 5
0.2 mm. c. A subgloboid spindle pole body with a flat internalized layer (white arrow) and radiating microtubules (black
arrows). The nucleus is indicated (N). Bar 50.2 mm. d. A mitochondrion (M) and a layered structure (white arrow head),
resembling a microscala. Bar 50.1 mm. e. Cell forming two buds at the same time. Note the thick layer of extracellular matrix
(asterisk). Bar 50.2 mm. f. Golgi complex (G) with numerous vesicles (white arrows) in the tip of a forming bud. Bar 50.2 mm.
g–h. Cconnection between the mother and daughter cell after the bud has fully matured. Bars 50.5 and 0.2 mm respectively. i.
Bud scar, showing the cell wall layers of the mother cell (black arrowheads) and a new cell wall formed after the daughter cell
detached. Bar 50.2 mm.
490 MYCOLOGIA
Meredithblackwellia eburnea Toome & Aime, sp. nov.
FIGS.1,2
MycoBank MB801223
Etymology: eburnea
, from Latin,
eburneus
, referring to the
cream (ivory, yellowish white) color of the colonies in culture.
Diagnosis: Meredithblackwellia eburnea
is similar to
Rhodotorula rosulata
Golubev & Scorzetti, differing in
that
M. eburnea
does not form a pseudomycelium and
is able to assimilate D-ribose and a-methyl-D-glucoside,
and unable to assimilate L-rhamnose, myo-inositol,
succinate and ethanol. While
M. eburnea
can grow in
the absence of vitamins,
R. rosulata
cannot.
Type:
GUYANA: Region 8, Potaro-Siparuni: Pakar-
aima Mountains, Upper Potaro River Basin, ,15 km
east of Mount Ayanganna, 5u18904.80N, 59u54940.40W;
710 m, from surface of undetermined fern leaf, 28
May 2010, leg. M.C. Aime, MCA4105 (NRRL Y-48821
– HOLOTYPE as lyophilized tissue; CBS 12589 and
ATCC MYA-4884 – ISOTYPES).
On YMA and PDA media, the colonies are smooth,
butyrous and creamy white with entire margins. Single
colonies are round with elevated center. After 3 d in
YM broth, cells are elongate, slightly fusiform, 3.9–5.2
312.6–17.6 mm (av. 4.7 314.8 mm), with width/length
ratio of 0.25/0.4 (av. 0.32). The cells contain several
vacuoles and lipid bodies (FIG. 2a), and are found
either singly or in rosettes (FIG. 1b, c). Budding is
enteroblastic and occurs on a narrow base from each
pole; up to three buds have been observed developing
from the same cell at the same time. Spindle pole
bodies are subgloboid with a flat internalized layer
(FIG. 2c, d). Optimal growth is at 20–25 C.
FIG. 3. Phylogenetic trees illustrating the position of
Meredithblackwellia eburnea
within Microbotryomycetes. a. Maximum
likelihood tree based on the sequences of LSU, SSU and ITS (first dataset), showing the position of
M. eburnea
in
Microbotryomycetes
and the five orders within it. The topology was rooted with
Mixia osmundae
. b. Maximum likelihood tree
based on LSU and ITS regions (second dataset) of Kriegeriales, showing the position of
M. eburnea
and composition of the
families Camptobasidiaceae and Kriegeriaceae. The topology was rooted with
Microbotryum violaceum
. On both trees the
numbers above the branch provide bootstrap (left) and maximum parsimony (right) values and the Bayesian posterior
probability value is given under the branch. GenBank accession numbers for used sequences are presented in SUPPLEMENTARY
TABLE I.
TOOME ET AL.: HIGHER-LEVEL CLASSIFICATION IN MICROBOTRYOMYCETES 491
Fermentation ability is negative. The following
carbon compounds are assimilated: D-glucose, D-
ribose, D-arabinose, sucrose, maltose, trehalose, a-
methyl-D-glucoside, cellobiose, salicin, arbutin, melez-
itose, glycerol, D-mannitol, D-glucono-1.5-lactone, DL-
lactate and quinic acid. Weak growth was detected on
D-galactose, L-sorbose, D-xylose, L-arabinose, L-rham-
nose, ribitol, xylitol and ethanol. No growth occurs on
D-glucosamine, melibiose, lactose, raffinose, inulin,
soluble starch, D-glucitol, galactitol, myo-inositol,
succinate, citrate, methanol or propane-1.2-diol.
Among nitrogen compounds, ethylamine is assimilat-
ed; weak growth was detected on L-lysine and D-
glucosamine. No growth occurs in the presence of
10%or 16%NaCl or 60%glucose; however, weak
growth was detected on 50%glucose. The culture
grows well in vitamin free media (some assimilation
results are in TABLE I).
Material examined: MCA4105
(NRRL Y-48821 5CBS
12589 5ATCC MYA-4884).
Camptobasidiaceae R.T. Moore
The family was established by R.T. Moore to
accommodate
Camptobasidium hydrophilum
(Moore
1996) and originally was placed in Atractiellales. Our
phylogenetic analysis shows that this family belongs to
Kriegeriales (FIG. 1B). Camptobasidiaceae contains
two known genera:
Camptobasidium
Marvanova´&
Suberkropp (Mycologia 82:209, 1990) and
Glaciozyma
Turchetti, Connell, Thomas-Hall & Boekhout (Ex-
tremophiles 15:579, 2011).
Known members of the Camptobasidiaceae are
C.
hydrophilum
,
G. antarctica
,
G. watsonii
,
G. martini
,
Leucosporidium
sp. AY30 and Antarctic yeast CBS8941.
DISCUSSION
The phylogenetic analyses of the first dataset of LSU,
SSU and ITS rDNA show that MCA4105 belongs to
Pucciniomycotina, Microbotryomycetes, forming a
supported ordinal clade, Kriegeriales ord. nov. with
TABLE I. Comparison of physiological characteristics of
Meredithblackwellia eburnea
and other members of Kriegeriales
1
a
2
b
3
c
4
d
5
e
6
f
7
g
8
h
9
i
10
j
Carbon source
D-Galactose 2
k
2++22 2 ++v
D-Ribose +2+222 2 2 v2
D-Xylose w w +w n/a 2n/a w/- v v
L-Arabinose 22 ++n/a 22w/- v 2
D-Arabinose +222222222
L-Rhamnose w ++2n/a 2+222
Melezitose ++++n/a +22 w2
Glycerol +w+222 2 v2+
myo-Inositol 2+2222 2 +22
DL-Lactate +dd222 2 n/a n/a n/a
Citrate 2d++22 2 vv2
Nitrogen source
Nitrate (K) 2+++++ + + + +
Nitrite (Na) 2++222 2 ww+
Ethylamine ++++++ + w w n/a
Others
50%D-Glucose w n/a 22222222
w/o vitamins +2+n/a n/a n/a n/a +++
Max growth T C 30 25 25 22 20 15 20 20 20 17
a
1.
M. eburnea
.
b
2.
Rhodotorula rosulata
(Golubev and Scorzetti 2010).
c
3.
Kriegeria eriophori
(Kurtzman et al. 2011).
d
4.
R. himalayensis
(Shivaji et al. 2008).
e
5.
R. glacialis
(Margesin et al. 2007).
f
6.
R. psychrophila
(Margesin et al. 2007).
g
7.
R. psychrophenolica
(Margesin et al. 2007).
h
8.
Glaciozyma martinii
(Turchetti et al. 2011).
i
9.
G. watsonii
(Turchetti et al. 2011).
j
10.
G. antarctica
(Kurtzman et al. 2011).
k
+5positive; 25negative; w 5weak positive; d 5delayed growth; v 5variable; n/a 5data not available.
492 MYCOLOGIA
the sedge parasite
Kriegeria eriophori
(FIG.3A).
Expanded sampling shows Kriegeriales comprise two
family clades, the previously described Camptobasi-
diaceae and Kriegeriaceae fam. nov. The family
Kriegeriaceae accommodates
K. eriophori
, a hyphal
fungus with a white yeast state (Double´s and
McLaughlin 1992), several anamorphic white yeasts
that have been isolated from plant surfaces from
temperate and tropical climates and glacier-associated
white yeasts. All these species form elongated yeast
cells with polar budding, which sometimes occurs in
small chains or rosettes. Camptobasidiaceae, original-
ly a monotypic family placed in Atractiellales (Moore
1996) but shown to belong to Microbotryomycetes
incertae sedis in Aime et al. (2006), is a supported
member of Kriegeriales (FIG. 3B). Camptobasidiaceae
contains a teleomorphic aquatic hyphal fungus
Camptobasidium hydrophilum
(Marvanova´ and Suber-
kropp 1990) and the recently described genus
Glaciozyma
, which contains psychrophilic white yeasts
isolated from Antarctic sea water or glacier sediments
(Turchetti et al. 2011). Although
Glaciozyma
species
grow mostly as yeast, they often also form hyphae and
teliospores. In addition to the taxa mentioned above,
several anamorphic white
Rhodotorula
spp. are refer-
able to Kriegeriales (FIG. 3B).
Morphologically
M. eburnea
resembles
R. rosulata
,
especially in the formation of rosettes in culture
(Golubev and Scorzetti 2010; FIG. 1c). Unlike
R.
rosulata
, however,
M. eburnea
has not been observed
to form pseudomycelium in culture. The two species
share 95%identity (527/556 bp) at the LSU and 88%
(410/464 bp) at the ITS, and phylogenetic analyses
show these species are clearly related and may even be
congeneric (FIG. 3B). However, we defer transferring
R. rosulata
to
Meredithblackwellia
in the absence of
strong molecular support (FIG. 3B) given that the aim
of this study was to describe
M. eburnea
and resolve
higher-level placement for several species previously
placed incertae sedis. Nonetheless, it is clear that in
the future the description of new genera in Krieger-
iales will be necessary for several species currently
placed in
Rhodotorula
s.l.
There exists little ultrastructural data for members
of Kriegeriales. However, the only species to have
been studied in detail,
K. eriophori
, has been shown to
produce subgloboid spindle pole bodies with a flat
internalized layer (Swann et al. 1999), which closely
resemble those observed in
M. eburnea
(FIG. 2c). This
spindle pole body type is also described as character-
istic for other Microbotryomycetes (Celio et al. 2006).
Although mitosis has been studied in a relatively small
number of basidiomycete yeasts (e.g. McCully and
Robinow 1972, McLaughlin et al. 1996), our study
confirms the present view that nuclear division in the
yeasts of Microbotryomycetes occurs in the bud
(FIG. 1f–k). One additional feature of note was the
presence of a layered structure in
M. eburnea
(FIG. 2d) that somewhat resembles the microscala
described in other select basidiomycete yeasts
(McLaughlin 1990). However, microscala have not
been found in any other species in Microbotryomy-
cetes and the microscala-like structures in
M. eburnea
seem to lack the cross connections between layers that
are characteristic for these structures.
The great variety of habitats from which the isolates
with the closest BLASTn matches to
M. eburnea
were
recovered suggests that Kriegeriales is an ecologically
diverse group of fungi. Based on the species known to
date, representatives of Camptobasidiaceae have been
isolated from water and glacier sediments. Krieger-
iaceae species, on the other hand, are mostly found in
association with plants in temperate climates but also
in association with glaciers. Therefore,
M. eburnea
,as
a fungus isolated in the Neotropics, represents a new
ecozone distribution for this group. Because habitats
such as ice, Antarctic sea, or leaf surfaces in tropical
areas are poorly studied for fungal diversity, it is
expected that further studies of these and other
undersampled habitats will increase species discovery
in Microbotryomycetes.
ACKNOWLEDGMENTS
This study was supported by Assembling the Fungal Tree of
Life (AFTOL) project NSF DEB-0732968. We thank Terry
Henkel for facilitating long term research in Guyana, Claire
Whittaker for general laboratory assistance and Tomas
Rush, Sebastian Albu, Gregory Heller and Jorge Diaz for
assistance with the assimilation experiments. We are
grateful for the help of David Lowry and Ricardo Reyes
from the School of Life Sciences Electron Microscope
Facility at Arizona State University with TEM and DAPI
staining sample preparation and David McLaughlin for his
expertise in TEM image interpretation. Anonymous review-
ers made useful suggestions on earlier versions of this
manuscript, and Else Vellinga provided advice on Latin.
This is number 196 in the Smithsonian’s Biological Diversity
of the Guiana Shield Program publication series.
LITERATURE CITED
Aime MC, Matheny PB, Henk DA, Frieders EM, Nilsson RH,
Piepenbring M, McLaughlin DJ, Szabo LJ, Begerow D,
Sampaio JP, Bauer R, Weiss M, Oberwinkler F, Hibbett
DS. 2006. An overview of the higher level classification
of Pucciniomycotina based on combined analyses of
nuclear large and small subunit rDNA sequences.
Mycologia 98:895–905, doi:10.3852/mycologia.98.6.896
———, Toome M, McLaughlin DJ. 2012. Pucciniomycotina.
In: McLaughlin DJ, Spatafora JW, eds. Mycota. Vol. VII,
2nd ed. Systematics and evolution (In press).
TOOME ET AL.: HIGHER-LEVEL CLASSIFICATION IN MICROBOTRYOMYCETES 493
Amend AS, Seifert KA, Bruns TD. 2010. Quantifying
microbial communities with 454 pyrosequencing: Does
read abundance count? Mol Ecol 19:5555–5565,
doi:10.1111/j.1365-294X.2010.04898.x
Andrews JH, Harris RF. 2000. The ecology and biogeogra-
phy of microorganisms on plant surfaces. Annu Rev
Phytopathol 38:145–180, doi:10.1146/annurev.phyto.
38.1.145
Bauer R, Begerow D, Oberwinkler F, Marvanova´ L. 2003.
Classicula
: the teleomorph of
Naiadella fluitans
.
Mycologia 95:756–764, doi:10.2307/3761949
———, ———, Sampaio JP, Weiss M, Oberwinkler F. 2006.
The simple-septate basidiomycetes: a synopsis. Mycol
Prog 5:41–66, doi:10.1007/s11557-006-0502-0
Begerow D, Bauer R, Oberwinkler F. 1997. Phylogenetic
studies on nuclear LSU rDNA sequences of smut fungi
and related taxa. Can J Bot 75:2045–2056, doi:10.1139/
b97-916
Buck JW. 2002. In vitro antagonism of
Botrytis cinerea
by
phylloplane yeasts. Can J Bot 80:885–891, doi:10.1139/
b02-078
Celio GJ, Padamsee M, Dentinger BTM, Bauer R, McLaugh-
lin DJ. 2006. Assembling the Fungal Tree of Life:
constructing the structural and biochemical database.
Mycologia 98:850–859, doi:10.3852/mycologia.98.6.850
Double´s JC, McLaughlin DJ. 1992. Basidial development,
life history and the anamorph of
Kriegeria eriophori
.
Mycologia 84:668–678, doi:10.2307/3760376
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-
Tallmann A. 2000. Biodiversity and systematics of
basidiomycetous yeasts as determined by large-subunit
rDNA D1/D2 domain sequence analysis. Int J Syst Evol
Microbiol 50:1351–1371, doi:10.1099/00207713-50-3-
1351
Gardes M, Bruns TD. 1993. ITS primers with enhanced
specificity for basidiomycetes: application to the iden-
tification of mycorrhiza and rusts. Mol Ecol 2:113–118,
doi:10.1111/j.1365-294X.1993.tb00005.x
Golubev WI, Scorzetti G. 2010.
Rhodotorula rosulata
sp. nov.,
Rhodotorula silvestris
sp. nov. and
Rhodotorula strami-
nea
sp. nov., novel
myo
-inositol-assimilating yeast
species in the Microbotryomycetes. Int J Syst Evol
Microbiol 60:2501–2506, doi:10.1099/ijs.0.016303-0
Hamamoto M, Nakase T. 2000. Phylogenetic analysis of the
ballistoconidium-forming yeast genus Sporobolomyces
based on 18S rDNA sequences. Int J Syst Evol Microbiol
50:1373–1380, doi:10.1099/00207713-50-3-1373
Hoch HC. 1986. Freeze substitution of fungi. In: Aldrich
HC, Todd WJ, eds. Ultrastructure techniques for
microorganisms. New York: Plenum Press. p 183–212.
Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008.
Dictionary of the Fungi. 10th ed. Wallingford, UK:
CABI.
Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: A
biologist-centric software for evolutionary analysis of
DNA and protein sequences. Briefings Bioinform 9:
299–306, doi:10.1093/bib/bbn017
Kumar TKA, Celio GJ, Matheny PB, McLaughlin DJ, Hibbett
DS, Manimohan P. 2007. Phylogenetic relationships of
Auriculoscypha
based on ultrastructural and molecular
studies. Mycol Res 111:268–274, doi:10.1016/j.
mycres.2006.12.003
Kurtzman CP, Fell JW, Boekhout T, eds. 2011. The yeasts. A
taxonomic study. 5th ed. Amsterdam: Elsevier.
Lee JK, Park KS, Park S, Park H, Song YH, Kang SH, Kim HJ.
2010. An extracellular ice-binding glycoprotein from
Arctic psychrophilic yeast. Cryobiology 60:222–228,
doi:10.1016/j.cryobiol.2010.01.002
Margesin R, Fonteyne PA, Schinner F, Sampaio JP. 2007.
Rhodotorula psychrophila
sp. nov.,
Rhodotorula psychro-
phenolica
sp. nov. and
Rhodotorula glacialis
sp. nov.,
novel psychrophilic basidiomycetous yeast species
isolated from alpine environments. Int J Syst Evol
Microbiol 57:2179–2184, doi:10.1099/ijs.0.65111-0
Marvanova´ L, Suberkropp K. 1990.
Camptobasidium hydro-
philum
and its anamorph,
Crucella subtilis
: a new
heterobasidiomycete from streams. Mycologia 82:208–
217, doi:10.2307/3759849
Matheny PB, Gossmann JA, Zalar P, Kumar TKA, Hibbett
DS. 2006. Resolving the phylogenetic position of the
Wallemiomycetes: an enigmatic major lineage of
Basidiomycota. Can J Bot 84:1794–1805, doi:10.1139/
b06-128
———, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH,
Hughes KW, Hofstetter V, Ammirati JF, Schoch CL,
Langer E, Langer G, McLaughlin DJ, Wilson AW,
Froslev T, Ge ZW, Kerrigan RW, Slot JC, Yang ZL,
Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT,
Vauras J, Hibbett DS. 2007. Contributions of rpb2 and
tef1 to the phylogeny of mushrooms and allies
(Basidiomycota, Fungi). Mol Phylogenet Evol 43:430–
451, doi:10.1016/j.ympev.2006.08.024
McCormack PJ, Wildman HG, Jeffries P. 1994. Production
of antibacterial compounds by phylloplane-inhabiting
yeasts and yeastlike fungi. Appl Environ Microbiol 60:
927–931.
McCully EK, Robinow CF. 1972. Mitosis in heterobasidio-
mycetous yeasts I.
Leucosporidium scottii
(
Candida
scottii
). J Cell Sci 10:857–881.
McLaughlin DJ. 1990. A new cytoplasmic structure in the
basidiomycete
Helicogloea
: the microscala. Exp Mycol
14:331–338, doi:10.1016/0147-5975(90)90056-Y
———, Frieders EM, Berres ME, Double´s JC, Wick SM.
1996. Immunofluorescence analysis of the microtubule
cytoskeleton in the yeast phase of the basidiomycetes
Kriegeria eriophori
and
Septobasidium carestianum
.
Mycologia 88:339–349, doi:10.2307/3760874
Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the
CIPRES science gateway for inference of large phylo-
genetic trees. New Orleans: Proceedings of the Gateway
Computing Environments Workshop. p 1–8.
Moore RT. 1996. An inventory of the phylum Ustomycota.
Mycotaxon 59:1–31.
Nakase T. 2000. Expanding world of ballistosporous yeasts:
distribution in the phyllosphere, systematics and
phylogeny. J Gen Appl Microbiol 46:189–216,
doi:10.2323/jgam.46.189
Nishida H, Ando K, Ando Y, Hirata A, Sugiyama J. 1995.
Mixia osmundae
: transfer from the Ascomycota to the
Basidiomycota based on evidence from molecules and
494 MYCOLOGIA
morphology. Can J Bot 73:S660–S666, doi:10.1139/
b95-308
Roberson RW, Fuller MS. 1988. Ultrastructural aspects of
the hyphal tip of
Sclerotium rolfsii
preserved by freeze
substitution. Protoplasma 146:143–149, doi:10.1007/
BF01405923
Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian
phylogenetic inference under mixed models. Bioinfor-
matics 19:1572–1574, doi:10.1093/bioinformatics/btg180
Sampaio JP. 2004. Diversity, phylogeny and classification of
basidiomycetous yeasts. In: Agerer R, Piepenbring M,
Blanz P, eds. Frontiers in basidiomycote mycology.
Eching, Germany: IHW-Verlag. p 49–80.
———, Gadanho M, Bauer R, Weiss M. 2003. Taxonomic
studies in the Microbotryomycetidae:
Leucosporidium
golubevii
sp. nov.,
Leucosporidiella
gen. nov. and the
new orders Leucosporidiales and Sporidiobolales.
Mycol Prog 2:53–68, doi:10.1007/s11557-006-0044-5
Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. 2002.
Systematics of basidiomycetous yeasts: a comparison of
large subunit D1/D2 and internal transcribed spacer
rDNA regions. FEMS Yeast Res 2:495–517.
Shivaji S, Bhadra B, Rao RS, Pradhan S. 2008.
Rhodotorula
himalayensis
sp. nov., a novel psychrophilic yeast
isolated from Roopkund Lake of the Himalayan
mountain ranges, India. Extremophiles 12:375–381,
doi:10.1007/s00792-008-0144-z
Swann EC, Frieders EM, McLaughlin DJ. 1999.
Microbotryum
,
Kriegeria
and the changing paradigm in basidiomycete
classification. Mycologia 91:51–66, doi:10.2307/3761193
———, Taylor JW. 1995. Phylogenetic perspectives on the
basidiomycete systematics: evidence from the 18S rRNA
gene. Can J Bot 73:S862–S868, doi:10.1139/b95-332
Swofford DL. 2002. PAUP* 4: phylogenetic analysis using
parsimony (*and other methods). Sunderland, Massa-
chusetts: Sinauer Associates.
Takashima M, Suh S-O, Nakase T. 1995. Phylogenetic
relationships among the species of the genus
Bensing-
tonia
and related taxa based on the small subunit
ribosomal DNA sequences. J Gen Appl Microbiol 41:
131–141, doi:10.2323/jgam.41.131
Turchetti B, Thomas Hall SR, Connell LB, Branda E,
Buzzini P, Theelen B, Mu¨ ller WH, Boekhout T. 2011.
Psychrophilic yeasts from Antarctica and European
glaciers: description of
Glaciozyma
gen. nov.,
Glacio-
zyma martini
sp. nov. and
Glaciozyma watsonii
sp. nov.
Extremophiles 15:573–586, doi:10.1007/s00792-011-
0388-x
Vilgalys R, Hester M. 1990. Rapid genetic identification and
mapping of enzymatically amplified ribosomal DNA from
several
Cryptococcus
species. J Bacteriol 172:4238–4246.
Whipps JM, Hand P, Pink D, Bending GD. 2008. Phyllo-
sphere microbiology with special reference to diversity
and plant genotype. J Appl Microbiol 105:1744–1755,
doi:10.1111/j.1365-2672.2008.03906.x
White TJ, Bruns TD, Lee S, Taylor JW. 1990. Amplification
and direct sequencing of fungal ribosomal RNA genes
for phylogenetics. In: Innis MA, Gelfand DH, Sninsky
JJ, White TJ, eds. PCR protocols: a guide to methods
and applications. Academic Press. p 315–322.
TOOME ET AL.: HIGHER-LEVEL CLASSIFICATION IN MICROBOTRYOMYCETES 495