Content uploaded by Akira Yokota
Author content
All content in this area was uploaded by Akira Yokota on Jul 30, 2017
Content may be subject to copyright.
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
INTERNATIONAL
JOURNAL
OF
SYSTEMATIC
BACTERIOLOGY,
July
1993,
p.
555-564
Copyright
0
1993, International Union
of
Microbiological Societies
OO20-7713/93/030555
-
10$02.00/0
Vol.
43,
No.
3
Proposal
of
Six
New Species
in
the
Genus
Aureobacterium
and Transfer
of
Flavobacterium esteraromaticum
Omelianski
to
the Genus
Aureobacterium
as
Aureobacterium
esteraromaticum
comb.
nov.
AKIRA
YOKOTA,’* MARIKO TAKEUCHI,’ TAKESHI SAKANE,l
AND
NOBERT
WEISS2
Institute
for
Fermentation, Osaka,
17-85,
Juso-honmachi 2-chome, Yodogawa-ku, Osaka
532,
Japan, and
Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH,
0-3300
Braunschweig, Germany2
Twelve strains placed in the genera
Flavobacterium, Pseudomonas,
and
Aureobacterium,
including soil
isolates, were characterized taxonomically.
On
the basis of morphological, physiological, and chemotaxonomic
data, as well as DNA-DNA hybridization data, we propose that 11
of
these strains should be classified in the
genus
Aureobacterium
as new combinations or new species,
as
follows:
Aureobacterium esteraromaticum
comb.
nov.
(type
strain,
IF0
3751
[=
ATCC
8091]),
Aureobacterium arabinogalacfanolyticum
sp. nov.
(type
strain,
IF0
14344),
Aureobacterium keratanolyticum
sp. nov. (type strain,
IF0
13309),
Aureobacterium luteolum
sp.
nov.
(type
strain,
IF0
15074
[
=
DSM
20143]),
Aureobucterium schleifen‘
sp. nov. (type strain,
IF0
15075
[
=
DSM
20489]),
Aureobucterium terrae
sp. nov.
(type
strain,
IF0
15300), and
Aureobacteriurn trichothecerwkjti-
cum
sp. nov. (type strain,
IF0
15077
[
=
JCM 13581). Whereas the peptidoglycan type
of
members
of
this genus
is considered to
be
B2f3, the
new
species
A. keratanolyticum
was shown to have a new peptidoglycan type,
murein variation
B2a
An
emended description of the genus
Aureobacterium
is presented.
The genus
Aureobacterium
was proposed by Collins et al.
(2), and the following
six
species have been described
previously
(8):
Aureobacterium liquefaciens, Aureobacte-
rium
jlavescens, Aureobacterium terregens, Aureobacte-
rium
saperdae, Aureobacteriurn barkeri,
and
Aureobacte-
rium testaceurn.
During the course of a taxonomic study of the
Flavobac-
terium
strains in the Institute for Fermentation at Osaka
(IFO) culture collection, we found that the aromatic com-
pound producers
Flavobacterium esteraromaticum
(18, 23)
and
“Flavobacterium suaveolens”
(24) and the keratan
sulfate endo-P-galactosidase (keratanase) producer
Pseudo-
monas
sp. strain
IF0
14344=
(T
=
type strain) (16, 17) were
members of the genus
Aureobacterium
(26).
To
determine
the taxonomic position of these organisms, we examined
their physiological and chemotaxonomic characteristics, as
well as the characteristics of some soil isolates and strains
designated
Aureobacterium
sp., and compared our results
with data for previously described species of the genus
Aureobacterium.
In this paper we describe the characteristics of 12 strains
of
Aureobacterium
species. We propose
six
new species and
one new combination in the genus
Aureobacterium
for these
strains on the basis
of
morphological, physiological, bio-
chemical, and DNA-DNA hybridization data.
MATERIALS
AND METHODS
Bacterial strains and culture conditions.
The bacterial
strains which we studied are listed in Table
1.
Biomass for
the chemical analyses was prepared by growing the strains at
28°C
with aerobic shaking in PY medium supplemented with
Bacto brain heart infusion (Difco Laboratories, Detroit,
Mich.) (PY-BHI medium), which contains
1%
peptone, 0.2%
yeast extract,
0.2%
brain heart infusion, 0.2% NaCl, and
*
Corresponding author.
555
0.2% D-glucose (pH
7.0).
Cells were harvested by centrifu-
gation, washed with water, and lyophilized.
Morphological, physiological, and biochemical characteris-
tics.
Unless otherwise indicated, all
of
the tests were carried
out at 28°C. Cell morphology was determined by examining
cells grown on PY-BHI agar. Motility was determined by the
hanging drop method. Catalase activity was determined by
bubbling in a 3% hydrogen peroxide solution. Oxidase
activity was determined by the oxidation
of
1% tetramethyl-
p-phenylenediamine on filter paper. Acid production from
carbohydrates was studied in a medium containing 0.3%
peptone,
0.25%
NaC1, 0.003% bromcresol purple, and
0.5%
carbohydrate (pH 7.2) (31). Assimilation of organic acids
was studied in a medium containing
0.5%
organic acid
(sodium salt),
0.02%
D-glucose,
0.01%
yeast extract, 0.01%
Trypticase (BBL), 0.1%
K2HP04,
0.5%
NaCl, 2% agar, and
12 mg
of
phenol red per liter (pH
7.0)
(31). Nitrate reduction
and hydrolysis of starch, gelatin, casein, and esculin were
tested by the methods described by Cowan and Steel (3).
Peptidoglycan analysis.
Cell walls were prepared from ca.
500
mg (dry weight)
of
cells by mechanical disruption with an
ultrasonic oscillator and were purified as described
by
Schlei-
fer and Kandler (22). The amino acid compositions of
complete wall hydrolysates were determined with a model
LC-6AD high-performance liquid chromatography (HPLC)
apparatus (Shimadzu
Co.,
Ltd., Kyoto, Japan) equipped
with a Wakopak WS-PTC column (Wako Pure Chemical
Industries, Ltd., Osaka, Japan) phenylthiocarbamoyl deriv-
atives according to the manufacturer’s instructions (29). The
amino acid compositions and isomers of diaminopimelic acid
were determined by developing preparations on cellulose
thin-layer chromatography plates
(Tokyo
Kasei Co., Ltd.,
Tokyo,
Japan), using two-dimensional descending chroma-
tography as described by Harper and Davis
(5).
The config-
urations of the amino acids were determined by measuring
the amino acid contents of the hydrolysates before and after
incubation with
D-
and L-amino acid oxidase (alanine and
homoserine), L-lysine decarboxylase (lysine), L-ornithine
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
556 YOKOTA ET
AL.
TABLE
1.
Bacterial strains
used
INT.
J.
SYST.
BACTERIOL.
Species Strain
(IF0
no.) Other designationu Reference(s)
or source Proposed reclassification
A. liquefaciens
A. flbvescens
A. terregens
A. saperdae
A. barkeri
A, testaceurn
F.
esterarornaticum
“F.
suaveolens”
Aureobacterium
sp.
Pseudomonas
sp.
Aureobacterium
sp.
Aureobacteriurn
sp.
Aureobacteriurn
sp.
Aureobacteriurn
sp.
Aureobacterium
sp.
Aureobacterium
sp.
Aureobacterium
sp.
Aureobacterium
sp.
15037T
15039T
12961T
15038T
15036T
12675T
3751T
3752
14344T
13309T
1530OT
15301
15302
15303
15077=
15074T
15075=
15076
ATCC 3647T
ATCC 13348T
ATCC 13345T
ATCC 19272=
ATCC 15954T
ATCC 15829T
ATCC 8091T
ATCC 958
JCM 1358T
DSM 20143=
DSM 20489*
DSM 20606
18
24
9
16, 17
Soil
Soil
Soil
Soil
15
27
21
21
A. esterarornaticurn
A.
esterarorna ticum
A.
ara binogalactanolyticurn
A. keratanolyticurn
A.
terrae
A. terrae
A. testaceurn
A. testaceurn
A.
trichothecenoiyticurn
A. luteolum
A. schleiferi
A. schleiferi
ATCC, American Type Culture Collection, Rockville, Md.; DSM, Deutsche Sammlung
von
Mikroorganismen und Zellkulturen GmbH, Braunschweig,
Germany; JCM, Japan Collection
of
Microorganisms, The Institute
of
Physical and Chemical Research
(RIKEN),
Wako,
Japan.
decarboxylase (ornithine), and L-glutamic acid decarboxy-
lase (glutamic acid) by using the method of Kandler and
Konig
(7).
Peptidoglycan structure was determined by the method of
Schleifer and Kandler (22). Partial acid hydrolysates were
subjected to by two-dimensional thin-layer chromatography,
and peptides were identified on the basis of their chromato-
graphic mobilities and staining characteristics (22). The
N-terminal amino acid of the interpeptide bridge was deter-
mined by dinitrophenylation of the undegraded peptidogly-
can (22).
Cell wall sugar analysis.
Cell walls were hydrolyzed with 2
N HC1 at
100°C
for 2 h, dried in vacuo, and then analyzed as
described by Mikami and Ishida (ll), using a model LC-5A
HPLC apparatus (Shimadzu Co., Ltd.) equipped with a
Shim-pack ISA 07/S2504 column (250 by 4 mm) and a
Shimadzu model RE-530 spectrofluorometer.
Glycolyl analysis.
Glycolate tests were performed as de-
scribed by Uchida and Aida (28).
Analysis
of
cellular fatty acids.
Fatty acids were extracted
from
50
mg (dry weight) cells by acid methanolysis and were
examined by using a model
GC-9A
gas-liquid chromatogra-
phy apparatus (Shimadzu Co., Ltd.) equipped with a glass
column (2 mm by
5
m) containing 10% diethyleneglycol
succinate on Chromosorb
W
at
180°C
(25).
Analysis of polar lipids.
Free lipids were extracted from
100 mg (dry weight) of cells, purified by the method of
Minnikin et al. (13), and examined by two-dimensional
thin-layer chromatography, using Kieselgel
60
FZs4
plates.
Lipids were visualized by spraying the plates with 10%
molybdophosphoric acid in ethanol and then heating them at
140°C for 10 min. The following specific spray reagents were
also used; a-naphthol for detection of sugars and ninhydrin
for detection of amino groups.
Analysis
of
mycolic acids.
Mycolic acids were analyzed by
the method of Minnikin et al. (12).
Analysis
of
isoprenoid
quinones.
Menaquinones were ex-
tracted from 200 mg (dry weight) of cells with chloroform-
methanol
(2:
1,
vol/vol), purified by thin-layer chromatogra-
phy in which benzene was used as the solvent, extracted
with diethyl ether, dried by using a nitrogen stream, and then
analyzed by HPLC, using a Shimadzu model LC-5A appa-
ratus equipped with a Zorbax octyldecyl silane column (4.6
by 150 mm).
DNA
base composition.
DNA was obtained by the method
of
Saito and Miura (19). The G+C content of the DNA was
determined by the method described by Mesbah et al. (10)
after treatment with P1 nuclease and alkaline phosphatase
and by HPLC by using a Shimadzu model LC-6AD appara-
tus equipped with a Comosil 5CI8-AR column (4.6 by 150
mm; Nacalai Tesque, Inc., Tokyo, Japan).
DNA-DNA
hybridization.
DNA-DNA hybridization was
performed fluorometrically in microdilution wells by using
biotinylated DNA (4).
RESULTS
AND DISCUSSION
Morphological, physiological, and biochemical characteris-
tics.
All of the strains studied were gram-positive rods that
were frequently arranged at an angle to give
V
forms. All
strains grew well on PY-BHI agar at
25
to
30”C,
and the color
of the colonies was yellow for all strains except IF0 14344T,
whose colonies were white. Among the 12 reclassified
strains, strains IF0 3751T, IF0 3752,
IF0
15302, and
IF0
15303 exhibited motility. The tests for which all 12 strains
gave positive results were the tests for catalase formation
and hydrolysis of esculin, while all 12 strains gave negative
results in oxidase and indole formation tests. None of the
strains except IF0 15302 and IF0 15303 produced acid from
the sugars examined (Table 2). Organic acids were utilized in
various combinations (Table 2). None of the strains required
terregens factor (2) for growth.
Chemical characteristics.
The chemical characteristics of
the test strains are summarized in Table 3. The amino acid
analysis and determination of the configurations of the amino
acids in the cell wall hydrolysates revealed the presence of
D-ornithine, D-alanine, D-glutamic acid plus hydroxyglu-
tamic acid, L-homoserine, and glycine (molar ratio, ca.
1:1:1:1:2) in all strains except strain IF0 13309T, whose
amino acids were D-ornithine, L-ornithine, malanine, D-glu-
tamic acid plus hydroxyglutamic acid, and glycine (molar
ratio, ca.
1:l:l:l:l).
Homoserine was not present. In addi-
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
c
r
0
P
w
-
dZff
-
+
-
dza
-
+
wx
dza
-
+
-
dza
-
+
PT-m
€1-XR
M
P
+
+
€;-XI4
Z1-XPI
+
+
+
+
+
+
+
M
+
-
q-XN
Zl-XN
+
+
+
+
+
+
+
M
+
M
Et-2IW
ZT-XN
+
+
+
+
M
+
+
P!-XW
ET-XW
M
M
M
M
ZT-XI4
‘11-XN
+
+
M
+
M
+
+
ZI-XPI
+
+
+
+
M
+
+
+
-
-
M
M
+
+
+
-
-
M
+
+
+
+
+
+
P
+
+
M
+
+
+
+
+
+
+
+
+
+
+
+
+
-
-
-
+
+
f
M
-
+
-
-
-
+
+
+
+
+
+
+
+
+
+ +
+
+
+
+
+
+
-
+
-
+ +
M
+
XO
-
+
+
XO
+
A
-
+
XO
+
h
-
A
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
558
YOKOTA
ET
AL.
INT.
J.
SYST.
BACTERIOL.
TABLE 3.
Chemotaxonomic characteristics
of
Aureobacterium
strainsa
Species Strain
(IF0
no.)
G+C
content
(mol%)
Major
menaquinone(s)
A.
liquefaciens
A.
flavescens
A.
terregens
A.
saperdae
A.
barkri
A.
testaceum
A.
testaceum
A.
testaceum
A.
esteraromaticum
A.
esteraromaticum
A.
arabinogalac-
tanolyticum
A.
kratanolyticum
A.
terrae
A.
terrae
A.
trichotheceno-
lyticum
A.
luteolum
A.
schleiferi
A.
schleifeii
15037T
15039=
12961T
15038T
15036=
12675T
15302
15303
3751T
3752
14344=
13309
1530OT
15301
15077T
15074T
15075T
15076
68.6
66.9
63.6
69.1
68.7
67.7
70.1
69.7
68.8
67.7
69.3
66.5
70.7
69.6
69.0
70.6
66.9
68.0
MK- 11,MK-12
MK- 13,MK-14
MK- 12, MK-
13
MK-ll,MK-12
MK-ll,MK-12
MK-
11
MK-11,MK- 10
MK-11
MK-12,MK- 13
MK-12,MK-13
MK-12,MK-13
MK- 12,MK-13
MK-
13, MK-
14
MK- 13, MK- 14
MK-12,MK-13
MK-12
MK-
1 1,
MK- 12
MK-
11,
MK-12,MK-10
Cell wall
sugar(s)d
Amino
acids in Polar lipids'
the cell wallb
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr, D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Orn,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
L-Hsr,D-Orn
+
D-Hyo
L-Hsr,D-Om +D-HYO
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
DPG, PG,
GL
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL,PGL
DPG,PG,GL
DPG,PG,GL,PGLs
DPG,PG,GL,PGLs
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
DPG,PG,GL
Rha
Rha,Gal,Glc
Rha,6dTal,Gal
Ga1,Glc
Rha,Gal,Glc
Rha,Man,Gal
Rha,Gal
Rha,Gal
Ga1,Glc
Ga1,Glc
Gal
Gal
Rha,Gal,Glc
Rha,Gal,Glc
Ga1,Glc
Rha,Gal
6dTal,Man,Gal
6dTal,Man,Gal
*
Data for the type strains
of
A.
liquefaciens,
A.
Pavescens,
A.
terregens,
A.
saperdae,
A.
barken,
and
A.
testaceum
are from reference
2.
The
major cellular
fatty acids of all strains are 12-methyltetradecanoic acid and 14-methylhexadecanoic acid. All strains contained cell wall glycolyl groups, and none contained
mycolic acids.
L-Hsr, L-homoserine; D-Orn, D-ornithine; D-HYO, D-hydroxyornithine.
DPG, diphosphatidylglycerol; PG, phosphatidylglycerol;
GL,
glycolipid; PGL, phosphoglycolipid; PGLs, phosphoglycolipids.
Rha, rhamnose; Gal, galactose; Glc, glucose; Man, mannose; 6dTa1, 6-deoxytalose.
tion, all strains except
IF0
3751T and
IF0
3752 contained a
small amount
of
L-lysine. Strains
IF0
15075T and
IF0
15076
contained equal amounts
of
D-ornithine and D-hydroxyorni-
thine, as noted previously by Schleifer et al. (21).
The peptidoglycan studies
of
Schleifer and Kandler (22)
showed that members of the genus
Aureobacterium
con-
tained a B2P type
of
peptidoglycan (Fig. 1B). All
of
the test
strains except strain
IF0
13309T contained the B2P type of
peptidoglycan; an examination
of
the primary structure
of
the murein of strain
IF0
13309T indicated that its peptidogly-
can type was B2a (Fig. 1A).
All strains contained high levels
of
glycolate in the glycan
moiety
of
the cell walls, which suggests that the muramic
acid occurs in the N-glycolyl form rather than the more
common N-acetyl form. The cell wall sugars rhamnose,
6-deoxytalose, mannose, galactose, and glucose were de-
tected in various combinations (Table
3).
All
of
the strains produced very similar fatty acid profiles
consisting primarily
of
anteiso-methyl branched acids,
an-
teiso-CIS:,, and anteiso-C,,,,. Mycolic acid was not present
in any
of
the strains. All
of
the strains contained long
unsaturated menaquinones (10 to 14 isoprene units) (Table
The polar lipids diphosphatidylglycerol and phosphatidyl-
glycerol and an unidentified glycolipid were detected in the
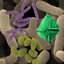
extracts
of
all strains. In addition, strains
IF0
14344*,
IF0
15300T, and
IF0
15301 contained more than one unidentified
glycolipid (Table
3
and Fig.
2).
All
of
the strains had DNA base compositions in the range
from 66.5 to 70.7 mol% G+C (Table 3).
On
the basis
of
chemical characteristics, all
of
the strains
except
IF0
13309T were identified as members
of
the genus
Aureobacterium
and could be differentiated from the genera
Curtobacterium
and
Microbacterium
and all other coryne-
form genera described previously (Table
5).
Strain
IF0
4)
-
13309T differed from the members
of
the genus
Aureobacte-
rium
in that it had a
B2a
type
of
peptidoglycan, but its
menaquinone composition, cellular fatty acid profile, and
phospholipid pattern and the presence
of
a glycolyl residue
in its muramic acid are typical
of
members
of
the genus
Aureobacteri~m~
This strain shows some resemblance to
members
of
the genus
Curtobacterium
since glycine is not
present in its cell wall but differs from
Curtobacterium
strains by its lack
of
L-homoserine, by its acyl type
of
muramic acid, and in its menaquinone system. Therefore,
the possibility that it belongs to the genus
Curtobacterium
can be excluded, and we concluded that strain
IF0
1330gT
should be included in the genus
Aureobacterium.
Hensel(6) described coryneform strain
S4
(=
DSM 20620)
which had a new murein type (type B2a); this organism is
somewhat similar to our strain
IF0
13309= but differs in
having glycine in the interpeptide bridge.
Our
results suggest
that strain S4 may also belong in the genus
Aureobacteriurn.
DNA-DNA
hybridization.
On
the basis
of
DNA-DNA
hybridization values (Table
6),
the test strains could
be
separated into seven distinct DNA relatedness groups. High
DNA relatedness values (>70%) were obtained between
IF0
3751T and
IF0
3752, between
IF0
15300 and
IF0
15301,
and between
IF0
15075T and
IF0
15076. Strains
IF0
15302
and
IF0
15303 produced high reassociation values with
A.
testaceum
IF0
12675T.
Species differentiation.
Differential characteristics for the
seven DNA relatedness groups and the
six
previously de-
scribed species
of
the genus
Aureobacterium
are summa-
rized in Table 2. Strains
IF0
15302 and
IF0
15303 had the
same phenotypic characteristics as
A. testaceum
and exhib-
ited high levels
of
DNA relatedness (Table 6) with that
species. Therefore, these strains were identified as members
of
A.
testaceum.
The cell walls
of
members of the genus
Aureobacterium
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
VOL.
43,
1993
TAXONOMY
OF
THE GENUS
AUREOBACTERZUM
559
TABLE
4.
Menaquinone compositions
of
the test strains
GlcNAc
-
MurNAc
-
GlcNAc
4
+
*
---,
Gly
Y
+
D-Ala
GlcNAc
-
MurNAc
-
GlcNAc
4
GlY
+
C
Glu -Gly
i
(Hyg)
;
L-Hsr
4
D-Ala
L-om
(Hyg)
D-Glu
Y
GIcNAc
-
-
L-Hsr
.)
FIG.
1.
Fragments
of
proposed primary structures
of
the pepti-
doglycans
ofA. kerutunolyticum
IF0
13309T
(A) and
A.
liquefaciens
(B).
The structure
of
A.
Ziquefuciens
peptidoglycan is based on the
data
of
Schleifer
(20).
Abbreviations: Gly, glycine; D-Glu,
D-glu-
tamic acid; D-Om, D-ornithine; D-Ma, D-alanine; L-Om, L-orni-
thine; Hyg, 3-hydroxyglutamic acid; L-Hsr, L-homoserine;
GlcNAc; N-acetylglucosamine; MurNAc, N-acetylmuramic acid.
The interpeptide bridge
is
enclosed by a dashed line.
contain a variety
of
sugars, including rhamnose, 6-deoxyta-
lose, mannose, galactose, and glucose (Table 3). The strains
in each DNA hybridization group contained the same com-
bination
of
sugars in their cell walls. Thus, the cell wall sugar
patterns
were characteristic for the species in this genus.
The genus
Aureobacterium
was established by Collins et
al.
(2).
It
is
characterized by the presence of D-ornithine in
the cell wall (B29 type of peptidoglycan) with a glycine in the
interpeptide bridge, N-glycolyl residues in the cell wall,
MK-11 to MK-14 isoprenoid quinones, G+C values of 66 to
69 mol%, and slow and weak production of acid from sugars.
On the basis of biochemical and chemical criteria, seven
DNA hybridization groups can be distinguished readily from
the previously described species
of
the genus
Aureobacte-
rium
and in our opinion warrant new taxa. Therefore, we
formally propose that the strains of these groups should be
classified in the genus
Aureobacterium
as follows:
Aureo-
bacterium esteraromaticum
comb. nov. for strains IF0
3751T
and
IF0
3752,
Aureobacterium arabinogalactanolyti-
cum sp.
nov. for strain IF0 14344T,
Aureobacterium
kera-
~~
%
of
menaquinones with
Strain the following no.
Species
(IFO
of isoprene units:
9
10 11
12
13 14
15
A.
liquefaciens
A.
flavescens
A.
terregens
A.
saperdae
A.
barkeri
A.
testaceum
A.
testaceum
A.
testaceum
A.
esteraromaticum
A.
esteraromaticum
A.
arabinogalactanolyticum
A.
keratanolyticum
A.
terrae
A.
terrae
A.
trichothecenolyticum
A.
luteolum
A.
schleiferi
A.
schleiferi
15037=
15039T
12961T
1503gT
15036T
12675T
15302
15303
3751T
3752
14344T
13309T
1530OT
15301
15077T
15074T
15075T
15076
15 40 43 2
7 40 48 5
4 34 49 12
5
28 60 7
9 51 38 2
8
13 63 15
1
20 71 9
16 72 12
1
61 39
6 58 36
2
7
60 30
1
8
8
41 42 2
7 64 28
1
5 56 37 3
23 60 17
19 64 17
10 56 32
20 60 20
tanolyticum
sp. nov. for strain IF0 13309T,
Aureobacterium
luteolum
sp. nov. for strain
IF0
15074T,
Aureobacterium
schleifen'
sp. nov. for strains IF0 15075T and IF0 15076,
Aureobacterium terrae
sp. nov. for strains IF0 15300T and
IF0
15301, and
Aureobacterium trichothecenolyticum
sp.
nov. for strain IF0
15077T.
An
emended description of the genus
Aureobacterium
and
descriptions of the new combination and new species are
given below.
Emended description of the genus
Aureobacterium
Collins,
Jones, Keddie, Kroppenstedt, and Schleifer.
Aureobacterium
(Aur.e.o.bac.te'ri.um.
L.
adj.
aureus,
golden; Gr. neut. n.
bakterion,
a small rod;
M.
L.
neut. n.
Aureobacterium,
small
golden rod). The salient characteristics of this genus, based
on the description of Collins et al.
(2)
and our own observa-
tions, are as follows. In young cultures, small irregular rods
occur; some cells are arranged at an angle, forming
V
shapes. In older cultures (ca. 3 to 7 days) the rods are
shorter, but a marked rod-coccus cycle (characteristic of the
genus
Arthrobacter)
is not observed. Primary branching
is
uncommon. The rods are gram positive and not acid fast;
metachromatic granules are not formed; endospores are not
formed. Growth occurs on suitable solid media in air.
Growth is inhibited by 6.5% NaC1. Growth occurs at ca. 10
to 40°C; the optimum temperature is ca.
25
to
30°C.
Obli-
gately aerobic. Slow and weak oxidative production of acid
from some carbohydrates occurs. Nutritionally exacting.
Some organic acids are utilized. Catalase positive. Oxidase
negative. Indole negative. Esculin
is
hydrolyzed. Noncellu-
lolytic.
The cell wall peptidoglycan is based on D-ornithine or
D-hydroxyornithine (type
B2P
of Schleifer and Kandler
[22])
([~-Hsr]-~-G1u-Gly-~-Orn)
or on
D-
and L-ornithine (type
B2a
of Schleifer and Kandler
[
221)
([~-0rn]-~-Glu-D-0rn).
The glycan moiety of peptidoglycan contains glycolyl and
acetyl residues. Contains no mycolic acids. The nonhydrox-
ylated fatty acids are predominantly anteiso- and iso-methyl
branched-chain acids; small amounts of straight-chain satu-
rated acids and traces of monounsaturated acids may be
present. Menaquinones are the sole respiratory quinones.
The major menaquinone isoprenologs contain various num-
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
560
YOKOTA ET
AL.
Im.
J.
SYST.
BACTERIOL.
FIG.
2.
Two-dimensional thin-layer chromatograms
of
olar lipids from
A.
Ziquefaciens
IF0
15037T (panel l),
A.
arabinogalactanolyticum
IF0
14344T (panel
2),
and
A.
keratanolyticum
IF0
13309
.p
(panel 3).
bers
of
isoprene units (10,
11,
12, 13, and 14 isoprene units).
The polar lipids are diphosphatidylglycerol, phosphatidyl-
glycerol, and unidentified glycolipids. DNA base composi-
tions range from 65 to 76 mol% G+C
(8).
The type species is
Aureobacterium liquefaciens.
Description
of
Aureobacterium esteraromaticum
(Omelian
ski
1923)
comb.
nov.
Aureobacterium esteraromaticum
(es. ter.
a.ro.ma‘ti.cum. German n.
ester,
ester; Gr. adj.
aromatikos,
sweet smelling; M.
L.
neut. adj.
esteraromaticum,
sweet
smelling because of esters). Gram-positive motile rods. The
motile cells have lateral flagella. Colonies are
1
to 3
mm
in
diameter, circular, low convex with entire margins, opaque,
and moist. A yellow pigment
is
produced. Many cells are
arranged at an angle, forming V shapes. The optimum
growth temperature is ca.
28°C.
No growth occurs in the
presence of 2.0% NaC1. The oxidation-fermentation test is
oxidative.
Acetate, malate, succinate, fumarate, lactate, propionate,
and hippurate are assimilated, but formate, citrate, and
oxalate are assimilated weakly or not assimilated. Starch,
Tween 40, Tween 60, and Tween
80
are hydrolyzed. Gelatin
is not hydrolyzed. Nitrate
is
not reduced. Voges-Proskauer
negative. Methyl red positive. Urease negative.
H,S
is
produced. Arginine dihydrolase is not produced.
The cell wall peptidoglycan contains ornithine, homo-
serine, glutamate plus hydroxyglutamate, glycine, and ala-
nine (molar ratio, ca. 1:1:1:2:1; variation B2P
of
Schleifer
and Kandler
[22]).
The cell wall sugars are galactose and
glucose. The major cellular fatty acids are anteiso- and
iso-methyl branched acids, anteiso-C,5:o, and anteiso-C,,:,.
Unsaturated menaquinones with 12 and
13
isoprene units are
present. The polar lipids are diphosphatidylglycerol, phos-
phatidylglycerol, and an unidentified glycolipid. The DNA
base composition
of
the type strain is 68.8 mol% G+C.
Source: soil
(18,
24).
The type strain is
IF0
3751
(=
ATCC 8091) (1, 18).
Flavobacten’um esteraromaticum
(18) is the basonym of
this species.
“Flavobacterium suaveolens”
IF0
3752 (=
ATCC
958)
(24, 30) is included in this species.
Description
of
Aureobacterium ara binogaluctanolyticum
s
p
.
nov.
Aureobacterium arabinogalactanolyticum
(a.ra.bi.no.
ga-lac. ta. no. ly
’
ti .cum. Engl. n.
arabinogalactan
,
poly-
saccharide produced by the bacterium
Mycobacterium tu-
berculosis;
Gr. adj
.
lyticum,
dissolving;
arabinogalac-
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
VOL. 43, 1993
TAXONOMY
OF
THE GENUS AUREOBACTERIUM
561
TABLE
5.
Distinguishing chemotaxonomic characteristics of coryneform taxa
Taxon Wall di- Murein
G+C
content
y2ir
Major menaquinone(s)
web
(mol%)
amino acid" Polar lipidsd,'
Agromyces L-DAB
B2y 71-76
Arcanobacterium LYS
A 48-52
Arthrobacter (globiformis L-LYS
A3a
59-69
Arthrobacter (nicotianae L-LYS
A4a 59-69
group)
group)
A
ureobacterium D-Om
B2P, B2a 67-70
Brachybacterium
Brevibacterium
Cellulomonas
Corynebacterium
Clavibacter
Curtobacterium
Exiguobacterium
Jonesia
Microbacterium
Nocardioides
meso-DAP
A4y 68-72
meso-DAP
Aly 60-67
meso-DAP
Aly 51-65
L-DAB
B2y 67-78
D-Om
B2P 68-75
LYS
A A 53-56
LYS
58-59
LL-DAP
A3y
69-72
L-Om
A4p, A4a 71-76
L-LYS
Bla, Blp 69-75
Terra bacter LL-DAP
A3y
69-72
Aeromicrobium LL-DAP
A3y
69-72
Ratobacter L-Om
A
65-66
Renibacterium L-LYS
A3a
52-54
Rubrobacter L-LYS
A3a
68
s,
A,
I
MK-11, MK-12, MK-13
DPG, PG, GL
s,
u
MK-9(H4)
ND
s,
A,
1
MK-9(H,)
DPG, PG, (PI), GL
s,
A,
I
MK-8, MK-9
DPG, PG, (PI), GL
MK-11, MK-12, MK-13,
MK-14
MK-7
MK-8(H2), MK-7(H,)
MK-9(H4)
MK-9(H2), MK-8(H2)
MK-10, MK-9
MK-9
MK-7
MK-9
MK-11, MK-12
DPG, PG, GL
DPG, PG,
GL
DPG, PG, (PI), GL
DPG, PI, PGL
DPG, PI, PIDM, (PG), (GL)
DPG, PG, GL
DPG, PG, GLs
DPG, PG, PE
DPG, PI, PGL
DPG, PG,
GL,
(PGL)
S,
A,
I,
U,
T,
MK-8(H4), MK-9(H4)
DPG, PG,
OH-PG
S,
A,
I,
U
MK-8(H4)
DPG,
PE,
PL, PGL
s,
u,
T None ND
s,
A,
I
MK-9
ND
s,
A,
I
MK-9, MK-10
DPG, GL
12-H,
A,
2-OH MK-8
DPG, PG, PL, PGL, GL
2-OH
a
L-DAB, L-diaminobutyric acid; Lys, lysine; L-LYS, L-lysine;
D-Om,
D-ornithhe;
meso-DAP,
meso-diaminopimelic acid; L-Om, L-ornithine; U-DN',
LL-diaminopimelic acid.
Murein type as described by Schleifer and Kandler (22).
S,
straight-chain saturated;
A,
anteiso-methyl branched;
I,
iso-methyl branched;
U,
monounsaturated;
T,
tuberculostearic acid; 12-H, 12-methylhexadecanoic
Parentheses indicate that a compound may or may not be present.
DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; GL, glycolipid;
PI,
phosphatidylinositol; PIDM, phosphatidylinositol dimannoside;
GLs,
glycolip-
ids; PE,
phosphatidylethanolamine;
OH-PG, 2-hydroxy fatty acid containing phosphatidylglycerol; PL, phospholipid; PGL, phosphoglycolipid; ND, not
determined.
acid;
2-0H,
2-hydroxylated fatty acids.
tanotyticum,
arabinogalactan dissolving.) Gram-positive,
nonmotile rods. Colonies are
1
to 3 mm in diameter, circular,
low convex with entire margins, opaque, and moist. No
pigment is produced. Many cells are arranged at an angle,
forming V shapes. The optimum growth temperature is ca.
28°C. Growth occurs in the presence
of
2.0% NaCl. The
oxidation-fermentation test is oxidative.
Acetate, malate, succinate, oxalate, fumarate, lactate,
propionate, and hippurate are assimilated, and formate and
citrate are weakly assimilated. Gelatin, starch, Tween 60,
and Tween 80 are hydrolyzed. Tween
20
and Tween 40 are
not hydrolyzed. Nitrate is not reduced. Voges-Proskauer
negative. Methyl red positive. Urease positive.
H,S
is pro-
duced. Arginine dihydrolase is produced.
The cell wall peptidoglycan contains ornithine, homo-
serine, glutamate plus hydroxyglutamate, glycine, alanine,
and a small amount
of
lysine (molar ratio, ca. 1:1:1:2:1:0.2;
variant B2P of Schleifer and Kandler [22]). The cell wall
sugar is galactose. The major cellular fatty acids are anteiso-
and iso-methyl branched acids, anteiso-C,,:,, and anteiso-
C17:o.
Unsaturated menaquinones with 12 and 13 isoprene
units are present. The polar lipids are diphosphatidylglyc-
erol, phosphatidylglycerol, an unidentified glycolipid, and
phosphoglycolipid. The G+C content
of
the DNA
of
the
type strain is 69.3 mol%. Produces a-D-arabinofuranosidase
(9,
14). Source:
soil
(9).
The type strain is
IF0
14344.
Description
of
Aureobucterium
kra&znolyticum
sp.
nov.
Aureobacterium keratanolyticum
(ker .a. ta.no. ly
'
ti-cum.
Engl. n.
keratan,
sulfur-containing polysaccharide produced
by mammals; Gr. adj.
lyticum,
dissolving;
keratanolyticum,
keratan dissolving). Gram-positive, motile rods. Colonies
are
1
to 3 mm in diameter, circular, low convex with entire
margins, opaque, and moist. A yellow pigment is produced.
Many cells are arranged at an angle, forming V shapes. The
optimum growth temperature is ca. 28°C. Growth occurs in
the presence of
2.0%
NaCl. The oxidation-fermentation test
is oxidative.
Acetate, lactate, malate, succinate, fumarate, propionate,
oxalate, and hippurate are assimilated, but citrate and for-
mate are assimilated weakly or not assimilated. Esculin and
gelatin are hydrolyzed. Starch, Tween 20, Tween 40, Tween
60, and Tween
80
are not hydrolyzed. Urease negative.
Nitrate
is
reduced. Voges-Proskauer negative. Methyl red
positive.
H,S
is produced. Arginine dihydrolase is produced.
The cell wall peptidoglycan contains ornithine but not
homoserine; the cell wall amino acids include ornithine,
glutamate plus hydroxyglutamate, glycine, alanine, and a
small amount
of
glycine (molar ratio, ca. 2:l:l:l:l:OS;
variation B2a
of
Schleifer and Kandler [22]). The cell wall
sugar is galactose. The major cellular fatty acids are anteiso-
and iso-methyl branched acids, anteiso-C,,:,, and anteiso-
C1,:o.
Unsaturated menaquinones with 12 and 13 isoprene
units are present. The polar lipids are diphosphatidylglyc-
erol, phosphatidylglycerol, and an unidentified glycolipid.
The G+C content of the DNA of the type strain
is
66.5
mol%. Produces keratan sulfate endo-P-galactosidase (EC
3.2.1.103) (16, 17). Source: soil (16).
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
INT.
J.
SYST.
BACTERIOL.
562
YOKOTA
ET
AL.
TABLE
6.
Levels
of
DNA
relatedness between test strains and the type strains
of
members
of
the genus
Aureobacterium
%
of
DNA-DNA
reassociation
with:
Species
A. liquefaciens
A. fravescens
A. terregens
A, saperdae
A.
barkri
A. testaceum
A. testaceurn
A. testaceum
A. esteraroma ticum
A. esteraromaticurn
A.
arabinogalactanotyticum
A. keratanolyticum
A. terrae
A. terrae
A.
trichothecenolyticum
A. luteolum
A. schleiferi
A. schleiferi
15037=
15039T
12961T
15038T
15036T
12675iT
15302
15303
375
lT
3752
14344T
13309T
153MT
15301
15077T
15074T
15075T
15076
100 9 13 16 14 16 22
5
17
1
12 2 12 10
11
16 13 9 9 19 18 22 18 10 8 26 10 12 8 23
28
100
38 19 21
5
15 20 20 4 13
5
10
11
2
2
9 15
11
29
11
25 8 17
3
5
22
13
7 3
1
18 100 17 17 6 23 19 13 24 14 3 10 7 10
3 33 41 100 87 13
11
8 12 4 14
8
8 9 8
16 20 19 75 92 28 10 17 17 3 16 7
11
8
13
1
21
8
87 100 15 17 14 10 2 13 21 12 8 7
5 33 7 2 9 100 99 10 21 8 7 13 10
6
2
4 13 8 3 6 96 100
11
14
4
6
11
32 6
0
7
9 12 10
1
20 3 100 32 9
11
4 10 33 7
13 29 3
5
10 17
1
5
100 15 7 5 17 25
5
7 16 23 3 16
15
13 26 12 100 89 13 16 10 13
30 25 59
11
10 28 15 24 14 84 100 17 22 9 15
2 37
13
9
6 18 13 8 9
5
14 100 9 12
5
1
37
2
11 1
22 4 18 8 6 6 8 100
8
10
0
30
0
5
1
7
3 15 16
3
1
4 10 100 90
123 6 7 3 712 917 3 7 31386100
The type strain is
IF0
13309.
Description
of
Aureobacterium terrae
sp.
nov.
Aureobacte-
rium terrae
(ter’rae.
L.
n.
terra,
soil;
M.
L.
gen. n.
terrae,
of
soil). Gram-positive, nonmotile rods. Colonies are
1
to 3 mm
in diameter, circular, low convex with entire margins,
opaque, and moist. A yellow pigment is produced. In young
cultures, small irregular rods are formed. Many cells are
arranged at an angle, forming V shapes. The optimum
growth temperature is ca. 28°C. Growth occurs in the
presence of 2.0% NaCl. The oxidation-fermentation test is
oxidative.
Acetate, fumarate, and lactate are assimilated, and for-
mate, citrate, malate, succinate, propionate, and hippurate
are assimilated weakly or not assimilated. Gelatin, starch,
Tween 40, Tween 60, and Tween
80
are hydrolyzed. Nitrate
is reduced. Voges-Proskauer and methyl red negative. Ure-
ase positive.
H,S
is formed. Arginine dihydrolase is not
produced.
The cell wall peptidoglycan contains ornithine, homo-
serine, glutamate plus hydroxyglutamate, glycine, alanine,
and a small amount of lysine (molar ratio, ca. 1:1:1:2:1:0.2;
variation
€32~~
of Schleifer and Kandler [22]). The cell wall
sugars are rhamnose, galactose, and glucose. The major
cellular fatty acids are anteiso- and iso-methyl branched
acids, anteiso-C,,,,, and anteiso-C,,:,. Unsaturated mena-
quinones with 13 and
14
isoprene units are present. The polar
lipids are diphosphatidylglycerol, phosphatidylglycerol, and
unidentified phosphoglycolipids. The
G+C
content of the
DNA
of the type strain
is
70.7
mol%. Source: soil.
The type strain is
IF0
15300.
Description
of
Aureobacterium trictaothecenolyticum
sp.
nov.
Aureobacterium trichothecenolyticum
(tri .cho
.
the. ce .no.
1y’ti.cum. Engl. n.
trichothecene,
a mycotoxin produced
by
the fungus
Trichothecium roseurn;
Gr. adj.
Zytzcum,
decom-
posing;
trichothecenolyticum,
trichothecene decomposing).
Gram-positive, nonmotile rods. Colonies are
1
to
3
mm in
diameter, circular, low convex with entire margins, opaque,
and moist. A yellow pigment is produced. Many cells are
arranged at an angle, forming V shapes. The optimum
growth temperature is ca. 28°C. Grows weakly in the pres-
ence of 2.0% NaCl. The oxidation-fermentation test
is
oxi-
dative.
Acetate, malate, succinate, and fumarate are assimilated;
formate, citrate, oxalate, lactate, propionate, and hippurate
are not assimilated. Starch, Tween 20, Tween 40, Tween
60,
and Tween 80 are hydrolyzed. Gelatin is not hydrolyzed.
Nitrate is reduced. Voges-Proskauer and methyl red nega-
tive. Urease positive.
H,S
is formed. Arginine dihydrolase is
not formed.
The cell wall peptidoglycan contains ornithine, homo-
serine, glutamate plus hydroxyglutamate, glycine, alanine,
and a small amount of lysine (molar ratio, ca. 1:1:1:2:1:0.4;
variation
B2a
of Schleifer and Kandler [22]). The cell wall
sugars are galactose and glucose. The major cellular fatty
acids are anteiso- and iso-methyl branched acids, anteiso-
C15:o, and anteiso-C,,:,. Unsaturated menaquinones with 12
and
13
isoprene units are present. The polar lipids are
diphosphatidylglycerol, phosphatidylglycerol, and an uniden-
tified glycolipid. The
G+C
content of the
DNA
of the type
strain is 69.0 mol%. Decomposes trichothecene mycotoxin
(8,
15).
Source: soil (15).
The type strain is
IF0
15077
(=
JCM
1358).
Description
of
Aureobacterium Cuteohm
sp.
nov.
Aureobac-
terium luteolum
(1u.te.o’lum.
L.
adj.
luteus,
yellow;
L.
neut.
adj
.
luteolum,
yellowish) Gram-positive, nonmotile rods.
Colonies are
I
to 3 mm in diameter, circular, low convex
with entire margins, opaque, and moist.
A
yellow pigment is
produced. Many cells are arranged at an angle, forming
V
shapes.
The
optimum growth temperature
is
ca.
28°C.
No
growth occurs in the presence
of
2.0% NaCl. The oxidation-
fermentation test is oxidative.
Acetate, malate, succinate, fumarate, lactate, propionate,
and hippurate are assimilated; formate, citrate, and oxalate
are assimilated weakly or not assimilated. Starch, Tween 20,
Tween 40, Tween
60,
Tween
80,
and gelatin are not hydro-
lyzed. Nitrate is reduced. Voges-Proskauer negative. Methyl
red positive. Urease positive.
H,S
is
not formed. Arginine
dihydrolase is not produced.
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
VOL. 43, 1993 TAXONOMY OF THE
GENUS
AUREOBACTERIUM
563
The cell wall peptidoglycan contains ornithine, homo-
serine, glutamate plus hydroxyglutamate, glycine, alanine,
and a small amount of lysine (molar ratio, ca. 1:1:1:2:1:0.5;
variation
B2a
of Schleifer and Kandler [22]). The cell wall
sugars are rhamnose, galactose, and glucose. The major cel-
lular fatty acids are anteiso- and iso-methyl branched acids,
antei~o-C~~:~, and anteiso-C,,,,. Unsaturated menaquinones
with 12 isoprene units are present. The polar lipids are
diphosphatidylglycerol, phosphatidylglycerol, and an uni-
dentified glycolipid. The G+C content
of
the DNA of the
type strain is 70.6 mol%. Source: soil (27).
The type strain is
IF0
15074
(=
DSM 20143).
Description
of
Aureobacterium
schleifri
sp.
nov.
Aureobac-
terium schleiferi
(sch1ei’fer.i.
L.
gen. n.
schleiferi,
of Schlei-
fer, referring to K.
H.
Schleifer, a German microbiologist
who contributed to the elucidation of the primary structure
of peptidoglycan and to taxonomic studies of the strains
belonging to this species). Gram-positive, nonmotile rods.
Colonies are
1
to 3 mm in diameter, circular, low convex
with entire margins, opaque, and moist.
A
yellow pigment is
produced. Many cells are arranged at an angle, forming V
shapes. The optimum growth temperature is ca. 28°C.
Growth occurs in the presence of 2.0% NaC1. The oxidation-
fermentation test is oxidative.
Organic acids are not assimilated. Tween 40 is hydro-
lyzed. Starch and gelatin are not hydrolyzed. Nitrate is not
reduced. Voges-Proskauer positive. Methyl red negative.
Urease negative.
H2S
is not produced. Arginine dihydrolase
is produced weakly.
The cell wall peptidoglycan contains ornithine plus hy-
droxyornithine, homoserine, glutamate plus hydroxygluta-
mate, glycine, alanine, and a small amount of lysine (molar
ratio, ca. 1:1:1:2:1:0.4 [21]; variation B2P of Schleifer and
Kandler [22]). The cell wall sugars are 6-deoxytalose, man-
nose, and galactose. The major cellular fatty acids are
anteiso- and iso-methyl branched acids, antei~o-C,~:~, and
anteiso-C,,:,. Unsaturated menaquinones with
11
and 12
isoprene units are present. The polar lipids are diphosphati-
dylglycerol, phosphatidylglycerol, and an unidentified glyco-
lipid. The G+C content of the DNA of the type strain is 66.9
mol%. Source: soil (21).
The type strain is
IF0
15075
(=
DSM 20489).
ACKNOWLEDGMENTS
We thank Kazuo Komagata,
Tokyo
University
of
Agriculture, for
valuable discussions and suggestions. We also thank Toru Hase-
gawa, Institute for Fermentation, Osaka, for support and discus-
sions.
1.
2.
3.
4.
5.
REFERENCES
Bergey, D. H.,
F.
C. Harrison,
R.
S.
Breed, B.
W.
Hammer, and
F.
M. Huntoon.
1930. Bergey’s manual
of
determinative bacte-
riology, 3rd ed. The Williams and Wilkins Co., Baltimore.
Collins, M. D.,
D.
Jones,
R.
M. Keddie,
R.
M. Kroppenstedt, and
K.
H. Schleifer.
1983. Classification
of
some coryneform bacte-
ria in a new genus,
Aureobacterium.
Syst. Appl. Microbiol.
k236-25 2.
Cowan,
S.
T., and K. J. Steel.
1965. Manual for the identification
of
medical bacteria. Cambridge University Press, London.
Ezaki,
T.,
Y. Hashimoto, and E. Yabuuchi.
1989. Fluorometric
deoqribonucleic acid-deoxyribonucleic acid hybridization in
microdilution wells as an alternative to membrane filter hybrid-
ization in which radioisotopes are used to determine genetic
relatedness among bacterial strains. Int. J. Syst. Bacteriol.
39:224-229.
Harper, J. J., and G. H.
G.
Davis.
1979. Two-dimensional
thin-layer chromatography for amino acid analysis
of
bacterial
cell walls. Int. J. Syst. Bacteriol. 29:56-58.
6.
Hensel,
R.
1984. Three new murein types in coryneform bacteria
isolated from activated sludge. Syst. Appl. Microbiol. 5:ll-19.
7.
Kandler,
O.,
and
H.
Konig.
1978. Chemical composition
of
the
peptidoglycan-free cell walls
of
methanogenic bacteria. Arch.
Microbiol. 118:141-152.
8.
Komagata, K., and
K.
Suzuki.
1986. Genus
Aureobacterium
Collins, Jones, Keddie, Kroppenstedt and Schleifer 1983,
672*, p. 1323-1325.
In
P. H. A. Sneath,
N.
S.
Mair, M.
E.
Sharpe, and J. G. Holt (ed.), Bergey’s manual
of
systematic
bacteriology, vol. 2. The Williams
&
Wilkins Co., Baltimore.
9.
Kotani,
S.,
T. Kato, T. Matsubara, M. Sakagoshi, and Y.
Hirachi.
1972. Inducible enzyme degrading serologically active
polysaccharides from mycobacterial and corynebacterial cells.
Biken J. 151-15.
10.
Mesbah, M., U. Premachandran, and
W.
B. Whitman.
1989.
Precise measurement of the G+C content
of
deoxyribonucleic
acid by high-performance liquid chromatography. Int.
J.
Syst.
Bacteriol. 39:159-167.
11.
Mikami, H., and Y. Ishida.
1983. Post-column fluorometric
12.
13.
14.
15.
16.
17.
18.
19.
20.
21.
22.
23.
24.
25.
26.
27.
detection of reducing sugars in high-performance liquid chroma-
tography using arginine. Bunseki Kagaku 32:E207-E210.
Minnikin,
D.
E.,
L.
Alshamaony, and M. Goodfellow.
1975.
Differentiation
of
Mycobacterium, Nocardia
and related taxa by
thin-layer chromatographic analysis of whole-organism metha-
nolysates. J. Gen. Microbiol. 88:200-206.
Minnikin, D.
E.,
M.
D.
Collins, and M. Goodfellow.
1979. Fatty
acid and polar lipid composition in the classification
of
Cellu-
lomonas, Oerskovia
and related taxa. J. Appl. Bacteriol. 4287-
95.
Misaki, A.,
I.
Azuma, and Y. Yamamura.
1977. Structural and
immunological studies on D-arabino-D-mannans and D-mannans
of
Mycobacterium tuberculosis
and other
Mycobacterium
spe-
cies. J. Biochem. 82:1759-1770.
Nakayama, K., A. Kato, Y. Ueno, Y. Minoda, and K. Komagata.
1980. Studies on the metabolism of trichothecene mycotoxins.
11.
Metabolism
of
T-2
toxin with the soil bacteria. Proc. Jpn.
Assoc.
Mycotoxicol. 12:30-32.
Nakazawa,
K.,
N.
Suzuki,
and
S.
Suzuki.
1975. Sequential
degradation
of
keratan sulfate by bacterial enzymes and purifi-
cation
of
a sulfatase in the enzymatic system. J. Biol. Chem.
Nakazawa, K., and
S.
Suzuki.
1975. Purification
of
keratan
sulfate-endogalactosidase
and its action on keratan sulfates
of
different origin. J. Biol. Chem. 250:912-917.
Omelianski,
V.
L.
1923. Aroma-producing microorganisms. J.
Bacteriol. 8393-419.
Saito, H., and
K.
Miura.
1963. Preparation
of
transforming
deoxyribonucleic acid by phenol treatment. Biochim. Biophys.
Acta 72:619-629.
Schleifer, K. H.
1970. Die Mureintypen in der Gattung
Mi-
crobacterium.
Arch. Mikrobiol. 71:271-282.
Schleifer, K. H., I. Hayn, H.
P.
Seidl, and
J.
Firl.
1983.
Threo-P-hydroxyornithine:
a natural constituent of the pepti-
doglycan
of
Corynebactenum
species Co 112. Arch. Microbiol.
Schleifer, K. H., and
0.
Kandler.
1972. Peptidoglycan types
of
bacterial cell walls and their taxonomic implications. Bacteriol.
Rev. 3k407-477.
Skerman,
V.
B.
D.,
V.
McGowan, and P. H.
A.
Sneath (ed.).
1980. Approved lists
of
bacterial names. Int. J. Syst. Bacteriol.
3a225-420.
Soppeland,
L.
1924.
Flavobacterium suaveolens,
a new species
of
aromatic bacillus isolated from dairy water.
J.
Agric. Res.
28:275-276.
Suzuki,
K., and K. Komagata.
1983. Taxonomic significance of
cellular fatty acid composition in some coryneform bacteria.
Int. J. Syst. Bacteriol. 33:188-200.
Takeuchi,
M.,
and
A.
Yokota.
1991. Reclassification
of
strains
of
Flavobactenum-Cytophaga
group in
IF0
Culture Collection.
Inst. Ferment. Res. Commun. (Osaka) 1583-96.
Topping,
L.
E.
1937. The predominant microorganisms in soils.
250905-911.
134:243-246.
Downloaded from www.microbiologyresearch.org by
IP: 191.96.247.155
On: Sun, 30 Jul 2017 18:10:32
564 YOKOTA ET
AL.
INT. J.
SYST.
BACTERIOL.
I. Description and classification
of
the organisms. Zentralbl.
Bakteriol. Parasitenkd. Infektionskr. Hyg. Abt. 2 97:289-304.
28. Uchida, K., and K. Ada. 1977. Acyl type
of
bacterial cell wall:
its simple identification by colorimetric method.
J.
Gen. Appl.
Microbiol. 23:249-260.
29. Wako
Pure
Chemical Industries, Ltd. 1989. Technical note on
the system
of
PTC-amino acid analysis. Wako Pure Chemical
Industries, Ltd., Osaka, Japan. (In Japanese.)
30. Weeks,
0.
B.
1974. Genus
Flavobactenum,
p. 357-364.
In
R.
E.
Buchanan and
N.
E.
Gibbons (ed.), Bergey’s manual
of
deter-
minative bacteriology, 8th ed. The Williams and Wilkins
Co.,
Baltimore.
31.
Yamada, K., and K. Komagata. 1972. Taxonomic studies on
coryneform bacteria.
IV.
Morphological, cultural, biochemical,
and physiological characteristics.
J.
Gen. Appl. Microbiol.
18:
3994 6.