Fig 3 - uploaded by Gert Van der Auwera
Content may be subject to copyright.
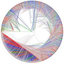
Unrooted phylogenetic tree for the Heterokonta based on a concatenated alignment of SSU and LSU rRNA sequences.
Source publication
The complete or nearly complete large-subunit rRNA (LSU rRNA) sequences were determined for representatives of several algal groups such as the chlorarachniophytes, cryptomonads, haptophytes, bacillariophytes, dictyochophytes and pelagophytes. Our aim was to study the phylogenetic position and relationships of the different groups of algae, and in...
Contexts in source publication
Context 1
... et al., , 1998Andersen et al., , 1999Potter et al., 1997 1997 ; Saunders et al., 1995Saunders et al., , 1997Bailey et al., 1998 ; Van de Peer & De Wachter, 1997a ; Van de Peer et al., 2000a). Therefore, we determined the complete or nearly complete LSU rRNA sequences of several species for which the SSU rRNA sequence was already known. Fig. 3 shows an unrooted phylogenetic tree of the Heterokonta (heterokont algae plus oomycetes, and hyphochytriomycetes) based on the concatenation of SSU rRNA and LSU rRNA sequences. The complete SSU rRNA sequence was used, except for the hypervariable region E23 (see Fig. 4), while three hypervariable regions of the LSU rRNA were omitted ...
Context 2
... Contrary to what has been previously suggested, the bacillariophytes do not form a monophyletic cluster with the pelagophytes and dictyochophytes, although they do share the possession of a reduced flagellar apparatus (Saunders et al., 1995). Maximum-likeli- hood analysis (not shown) shows the same topology and similar support as the tree in Fig. 3. Maximum- parsimony supports all the sister-relationships dis- cussed above, but support is lacking for any of the relationships between classes of heterokont algae at a deeper ...
Similar publications
Ribosomal RNA (rRNA) genes are widely utilized in depicting organismal diversity and distribution in a wide range of environments. Although a few cases of lateral transfer of rRNA genes between closely related prokaryotes have been reported, it remains to be reported from eukaryotes. Here, we report the first case of lateral transfer of eukaryotic...
A recent reclassification of diatoms based on phylogenies recovered using the nuclear-encoded SSU rRNA gene contains three major classes, Coscinodiscophyceae, Mediophyceae and the Bacillariophyceae (the CMB hypothesis). We evaluated this with a sequence alignment of 1336 protist and heterokont algae SSU rRNAs, which includes 673 diatoms. Sequences...
The East China Sea (ECS) around Jeju Island lies in a transitional region between the temperate and subtropical zones and is influenced by the Kuroshio Current. Hence, this area has been considered an important monitoring site for oceanic ecosystem changes. Herein, we assessed the community structure and diversity of the phytoplankton in the ECS ar...
A new marine pelagophyte genus, Andersenia, was described from clonal cultures established from sand samples collected in coastal tide pools from southeastern Australia. Thin, uniseriate, unbranched filaments were variable in length up to 1.0 mm, 7–12 μm in width and surrounded by a gelatinous envelope. Cells contained one or two golden brown, lobe...
Figure S1 depicts phylogenetic trees of fungal sequences, rooted with an oomycete (heterokont) outgroup. Figure S1a shows phylogenetic trees based on independent and combined analyses of ribosomal RNA and RPB2 gene loci, which reflect the organismal phylogeny. Figure S1b presents phylogenetic trees based on independent and combined analyses of gene...
Citations
... The secondary structures of the nuclear SSU and LSU rRNA genes were aligned based on secondary structures of the nr SSU and LSU rRNA gene sequences of the dictyochophycean species Apendinella radians (Ali et al., 2001). For species delimitation among the Dinobryon species, the E23-5 helix of the V4 region in nr SSU rRNA and the E11-1 helix of D7b and the E20-5 helix of the D8 regions in nr LSU rRNA were selected as molecular signatures. ...
Introduction
The genus Dinobryon is one of the most recognizable chrysophyte genera, characterized by dendroid colonies with a biflagellate inside each cellulosic lorica. The representative forms of lorica are cylindrical, conical, vase, or funnel shaped, with undulation on the lorica wall. Traditionally, the morphological characteristics of the lorica and the colony organization have been used for the delimitation of Dinobryon species.
Methods
To understand the taxonomy and phylogeny of colonial Dinobryon species, we performed molecular and morphological studies using 39 unialgal cultures and 46 single colony isolations from environmental specimens collected in Korea. We used a nuclear internal transcript spacer (ITS1-5.8S-ITS2) to find the genetic diversity of Dinobryon from environmental samples and a combined dataset from six gene sequences (nuclear SSU and LSU rRNA, plastid LSU rRNA, rbcL and psaA, and mitochondrial CO1 genes) for phylogenetic analysis.
Results and discussion
We found 15 different lineages based on the genetic diversity of the nuclear ITS sequences. The phylogenetic tree of the colonial species based on the combined multigene dataset were divided into 18 subclades, including five new species, each with unique molecular signatures for the E23-5 helix of the V4 region in the nuclear SSU rRNA and the E11-1 helix of D7b, and the E20-1 helix of D8 regions in the nuclear LSU rRNA. Morphological studies were focused on lorica dimension and shape, and stomatocyst morphology. The Dinobryon species showed similarities or differences in lorica morphologies between and within species, and also differences in lorica size between culture and environmental samples. Five Dinobryon species formed distinctive stomatocysts, their stomatocyst morphologies, including collar structure, surface ornamentation, and cyst shape, showed unique characteristics in each species and were useful for identification. Here, we propose five new species based on morphological and molecular evidences: D. cylindricollarium, D. exstoundulatum, D. inclinatum, D. similis, and D. spinum.
... Common molecular markers including 18S rDNA, 16S rDNA, ITS and rbcL have been used to study R. setigera phylogenetics (Ben Ali et al. 2001;Yu et al. 2018;Yoshimatsu et al. 2020). Although these molecular markers can be effectively used to differentiate R. setigera from other Rhizosolenia species, they are inadequate for distinguishing intraspecies strains. ...
... Despite its worldwide distribution, its genetic diversity that may have dictated its local adaptation has never been explored. Previous identification of R. setigera was mostly limited to the morphological description with limited usage of some common markers (Ben Ali et al. 2001;Yu et al. 2018;Yoshimatsu et al. 2020). However, morphological features are not enough to distinguish R. setigera strains. ...
Rhizosolenia setigera is cosmopolitan diatom with broad distribution in many ocean regions with diverse environmental features, suggesting that R. setigera may have rich genetic diversity. Because genetic diversity of many phytoplankton species is high, we hypothesize that R. setigera genetic diversity is also high and genetic diversity plays a role in its adaptation and bloom development although no evidence has been documented so far to suggest that R. setigera HAB events are a strain-specific phenomenon. Because common molecular markers including 18S rDNA, 16S rDNA, ITS, and rbcL can not resolve R. setigera genetic diversity due to limited resolution, high-resolution molecular markers are needed for resolving and tracking R. setigera genetic diversity and biogeographic distribution. Here, we designed a new molecular marker R. setigera mitochondrial 1 (rsmt1) with high-resolution and high-specificity by comparative analysis of mitochondrial genomes (mtDNAs) of R. setigera strains isolated from various coastal regions of China. We ascertained R. setigera genetic diversity in coastal regions of China using this newly developed molecular marker and identified distribution patterns of R. setigera strains, demonstrating that rsmt1 could be generally applied to probe R. setigera genetic diversity.
... The secondary structures of the nuclear (nr) SSU and LSU rRNA genes were aligned manually, with the alignment based on secondary structures of the nr SSU and LSU rRNA gene sequences of the dictyochophycean species Apendinella radians (Ali et al., 2001) as a guide in the Macgde (2.6) (Smith et al., 1994). As a molecular signature for Spumella-like flagellates, domain 8 from the V1 region of the nr SSU rRNA and D5 domain from the nr LSU rRNA gene were selected for identification of the genus Spumella. ...
... In particular, the V4 region of nr SSU rRNA is known as a useful genetic marker for understanding biological diversity in various protistan groups, including chrysophytes (Zimmermann et al., 2011;Bock et al., 2017;Tragin et al., 2017). The V4 region is also known as a hypervariable region (Ali et al., 2001;Tragin et al., 2017). The sequences in the molecular signature region are well conserved within species but variable among species in the genus Spumella. ...
The genus Spumella, established by Cienkowsky in 1870, is characterized by omnivory, two (rarely three) flagella, a short stick-like structure beneath the flagella, a threadlike stalk, cell division via constriction and cyst formation. Since the first phylogenetic study of Spumella-like flagellates, their paraphyly has consistently been shown, with separation into several genera. More recently, Spumella was carefully investigated using molecular and morphological data to propose seven new species. Classification of this genus and knowledge of its species diversity remain limited because Spumella-like flagellates are extremely difficult to identify based on limited morphological characters. To understand the phylogeny and taxonomy of Spumella, we analyzed molecular and morphological data from 47 strains, including 18 strains isolated from Korean ponds or swamps. Nuclear SSU, ITS and LSU rDNA data were used for maximum likelihood and Bayesian analyses. The molecular data divided the strains into 15 clades, including seven new lineages, each with unique molecular signatures for nuclear SSU rRNA from the E23-2 to E23-5 domains, the spacer between the E23-8 and E23-9 domains of the V4 region and domain 29 of the V5 region. Our results revealed increased species diversity in Spumella. In contrast to the molecular phylogeny results, the taxa showed very similar cell morphologies, suggesting morphological convergence into simple nanoflagellates to enable heterotrophy. Three new species produced stomatocysts in culture. Aspects of stomatocyst morphology, including collar structure, surface ornamentation, and cyst shape, were very useful in differentiating the three species. The general ultrastructure of Spumella bureschii strain Baekdongje012018B8 and S. benthica strain Hwarim032418A5 showed the typical chrysophyte form for the leucoplast, a vestigial chloroplast surrounded by four envelope membranes, supporting the hypothesis that Spumella evolved from a phototroph to a heterotroph via the loss of its photosynthetic ability. Seven new species are proposed: S. benthica, S. communis, S. longicolla, S. oblata, S. rotundata, S. similis, and S. sinechrysos.
... This was done to ensure including a representative sample of the diatom diversity for estimating our molecular clock rates. Overall, we could compile 18S, 28S and rbcl sequences from 76 diatom species encompassing the major groups [20][21][22][23][24][25][26][27][28][29][30][31][32][33] . There were too few COX-1 sequences available for these taxa and the available ITS and 5.8S sequences could not be aligned confidently, so these loci were not used in the dating analyses. ...
The diatom Didymosphenia geminata has gained notoriety due to the massive growths which have occurred in recent decades in temperate regions. Different explanations have been proposed for this phenomenon, including the emergence of new invasive strains, human dispersion and climate change. Despite the fact in Argentina nuisance growths began in about 2010, historical records suggest that the alga was already present before that date. In addition, preliminary genetic data revealed too high a diversity to be explained by a recent invasion. Here, we estimate the divergence times of strains from southern Argentina. We integrate new genetic data and secondary, fossil and geological calibrations into a Penalized Likelihood model used to infer 18,630 plausible chronograms. These indicate that radiation of the lineages in Argentina began during or before the Pleistocene, which is hard to reconcile with the hypothesis that a new variant is responsible for the local mass growths. Instead, this suggests that important features of present distribution could be the result of multiple recent colonizations or the expansion of formerly rare populations. The text explains how these two possibilities are compatible with the hypothesis that recent nuisance blooms may be a consequence of climate change.
... MG782510) and the sequence in GenBank (Accession no. AF289042), the source of which, however, could not be tracked from the submission information and the corresponding publication(Ben Ali et al., 2001). These results suggest that all populations of A. anophagefferens from China and USA, and that represented by the entity AF289042, share a fairly recent common irradiating origin, which cannot be identified with highly limited molecular data currently available. ...
The nonmotile, spherical, picoplanktonic (2‐μm‐sized) pelagophyte Aureococcus anophagefferens has caused numerous harmful blooms (“brown tides”) across global marine ecosystems. Blooms have developed along the east coast of the USA since 1985, a limited number of times in South Africa around 1997, and frequently in China since 2009. As a consequence, the harmful blooms have caused massive losses in aquaculture and coastal ecosystems, particularly mortalities in cultured shellfish. Therefore, whether A. anophagefferens was recently introduced to China via natural/artificial transport of resting stage cells or has been an indigenous species has become a question of profound ecological significance and broad interest, which motivated our extensive investigation on the geographic and historical presence of this species in the seas of China. We applied a combined approach of extensive PCR‐based detection and sequencing, germination experiments and monoclonal antibody staining of germlings to samples of surface sediment and sediment core (dated via combined isotopic measurements) collected from all four seas of China, and searched the supplementary data set of a recent Science publication. We discovered that A. anophagefferens does have a resting stage in the sediment, but it also has a wide geographic distribution both in China (covering a range of ~30° in latitude, ~15.7° in longitude and 2.5–3,456 m in water depth; temperate to tropical and coastal to open oceans) and in almost all oceans of the world and a historical presence of >1,500 years in the Bohai Sea, China. The work revealed that A. anophagefferens is not a recently introduced, but an indigenous species in China and has in fact a globally cosmopolitan distribution.
... Molecular-based analysis on phylogeny has started on a new page of evolutionary relationship and substituted the previous morphological-based classification. Several markers, for example, small subunit (SSU) rDNA [31], large subunit (LSU) rDNA [41], have been established to study algal phylogeny and evolution. In addition, the detail inheritance may be accomplished by developing specific protein coding genes, such as rbcL and matK in chloroplast [42,43], internal transcribed spacer (ITS) [35], and nrDNA [44]. ...
... Moreover, NGS technology has advanced a genomics approach to differentiate more precisely among orthologous and paralogous regions at different loci within different species. The obstacle that scientists faced was that the short sequences may not support to all branches in a phylogeny [41]. Two-locus barcoding with rbcL and matK instead of single marker analyses has improved the accuracy and resolution of phylogenetic reconstruction [46]. ...
Green algae, Chlorella ellipsoidea, Haematococcus pluvialis and Aegagropila linnaei (Phylum Chlorophyta) were simultaneously decoded by a genomic skimming approach within 18-5.8-28S rRNA region. Whole genomic DNAs were isolated from green algae and directly subjected to low coverage genome skimming sequencing. After de novo assembly and mapping, the size of complete 18-5.8-28S rRNA repeated units for three green algae were ranged from 5785 to 6028 bp, which showed high nucleotide diversity (π is around 0.5–0.6) within ITS1 and ITS2 (Internal Transcribed Spacer) regions. Previously, the evolutional diversity of algae has been difficult to decode due to the inability design universal primers that amplify specific marker genes across diverse algal species. In this study, our method provided a rapid and universal approach to decode the 18-5.8-28S rRNA repeat unit in three green algal species. In addition, the completely sequenced 18-5.8-28S rRNA repeated units provided a solid nuclear marker for phylogenetic and evolutionary analysis for green algae for the first time.
... Estos organismos poseen pigmentos fotosintéticos primarios como la clorofila a, otros pigmentos como los Beta-caroteno y fucoxantina (carotenoides), así como pigmentos accesorios: ficocianina y ficoeritrina (Martínez-Silva, 2010). De acuerdo a la presencia o ausencia de pigmentos, las microalgas pueden ser clasificadas en tres grandes grupos: aquellos que presentan clorofila a y b (algas verdes/Chlorophytes), clorofila a y ficobilinas (algas rojas/Rhodophytes), clorofila a y c (algas pardas-amarillas/Chromophytes), éstas últimas al ser un grupo polifilético se encuentra subdividido en cuatro taxones: Haptophytas, Dinoflagelados, Criptophytas y algas heterocontas o Xanthophytas (Ben Ali et al., 2001)Martínez-Silva, 2010). ...
... Previously unknown chloroplastic TG synthesis was discovered in three Chlamydomonas species besides C. reinhardtii and four other closely related algae. All of the algal species analyzed segregated into two distinct groups based on their %16/2 values (Fig. 5) coinciding with the evolutionary divergence of the order Chlamydomonadales [41]. This finding has important implications for the use of C. reinhardtii as a model species of TG analysis and demonstrates the utility of a high throughput TG quantification method. ...
A method for the specific quantification of triacylglycerides (TGs) using electrospray ionization (ESI) reverse-phase liquid chromatography tandem mass spectroscopy (LC–MS/MS) with multiple reaction monitoring (MRM) was developed for routine high throughput applications. Incorporation of catalytic hydrogenation dramatically reduced TG pool complexity, enabling the absolute quantification of TG and regioisomers at the expense of double bond frequency and placement. Saturated TAG standards demonstrated equivalent linear detector responses to increases in concentration over two orders of magnitude. Accuracy was evaluated against a standard gas chromatography mass spectroscopy (GC–MS) protocol using three plant-derived oils and biological lipid extracts of two algal genera, Chlamydomonas reinhardtii and Coccomyxa subellipsoidea. Quantification of TG in palm and olive oils was > 91% accurate compared to the GC–MS method, and the content of TG molecular species was comparable to literature values. Differences in algal TG quantities were statistically insignificant. The ratio of the product ion peak areas for pairs of regioisomers was linearly correlated to their relative abundance. Quantification of regioisomers enabled sn-2 positional analyses related to the amount of TG synthesized in the chloroplast of photosynthetic organisms. A demonstrative survey of 35 algae revealed an evolutionary divergence of the dominant TG synthetic pathways by order Chlamydomnadales.
... Phaeophytes are believed to have arisen with diatoms and golden algae (all belonging to Ochrophyta), as well as Oomycetes via the secondary endosymbiotic event (Reyes-Prieto et al., 2007). These organisms all belong to heterokonts, which are characterized by the occurrence of cells with two unequal flagella in their life history (Ben Ali et al., 2001). There continues to be heated debate over the origin of eukaryotic algae as monophyletic or polyphyletic. ...
Most phaeophytes (brown algae) and rhodophytes (red algae) dwell exclusively in marine habitats and play important roles in marine ecology and biodiversity. Many of these brown and red algae are also important resources for industries such as food, medicine and materials due to their unique metabolisms and metabolites. However, many fundamental questions surrounding their origins, early diversification, taxonomy, and special metabolisms remain unsolved because of poor molecular bases in brown and red algal study. As part of the 1 000 Plant Project, the marine macroalgal transcriptomes of 19 Phaeophyceae species and 21 Rhodophyta species from China’s coast were sequenced, covering a total of 2 phyla, 3 classes, 11 orders, and 19 families. An average of 2 Gb per sample and a total 87.3 Gb of RNA-seq raw data were generated. Approximately 15 000 to 25 000 unigenes for each brown algal sample and 5 000 to 10 000 unigenes for each red algal sample were annotated and analyzed. The annotation results showed obvious differences in gene expression and genome characteristics between red algae and brown algae; these differences could even be seen between multicellular and unicellular red algae. The results elucidate some fundamental questions about the phylogenetic taxonomy within phaeophytes and rhodophytes, and also reveal many novel metabolic pathways. These pathways include algal CO2 fixation and particular carbohydrate metabolisms, and related gene/gene family characteristics and evolution in brown and red algae. These findings build on known algal genetic information and significantly improve our understanding of algal biology, biodiversity, evolution, and potential utilization of these marine algae.
... The analyses presented here show that the raphidophytes included form a monophyletic group with high bootstrap support that includes both the marine and freshwater Vacuolaria genera. This agrees with previous studies (Potter et al., 1997;Ben Ali et al., 2001;Ben Ali et al., 2002;Hosoi-Tanabe et al., 2006). Members of the 4 different genera are quite distinct from each other. ...
The morphology and genetic affinity of a novel raphidophyte belonging to the genus Chattonella Biecheler is described for the first time from the Oman Sea along the south-east coast of Iran. While morphologically very similar to Chattonella subsalsa Biecheler, the Iranian isolates contain a distinct red eyespot. A comparison of LSU-rDNA and rDNA-ITS show that the Iranian isolate is genetically distinct from other Chattonella subsalsa strains isolated from across a wide global range and indicates that the Iranian isolates represent a distinct species related to Chattonella subsalsa.