Fig 2 - uploaded by Alma L Burlingame
Content may be subject to copyright.
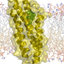
Structure of Mediator Head module – CTD complex. ( A ) Secondary structure representation of the Head module (green) superimposed on weighted electron-density map of the Head alone (2F O -F C ; blue mesh), contoured at 1.25 σ . Difference map showing electron density for the CTD (brown mesh), calculated with F soak -F native amplitudes and MIRAS phases, contoured at 2.5 σ . CTD model encompassing almost four heptapeptide repeats is shown in stick representation (yellow). ( B ) Same as A rotated 90 ° around the vertical axis. ( C ) Close-up view of CTD interaction region bounded by dashed box in B . ( D ) CTD model and density from C shown with an aligned section of unmodi fi ed CTD peptide ( β -turn ) from high-resolution crystal structure (PID: 3D9O).
Source publication
The X-ray crystal structure of the Head module, one-third of the Mediator of transcriptional regulation, has been determined as a complex with the C-terminal domain (CTD) of RNA polymerase II. The structure reveals multiple points of interaction with an extended conformation of the CTD; it suggests a basis for regulation by phosphorylation of the C...
Contexts in source publication
Context 1
... Mobile Jaw domains. Substructure refinement with the use of MIRAS phases resulted in electron density maps revealing many features not seen previously, such as domain connectivity in the central "Joint" region ( Fig. 1) and large sections of β-sheet within both the Med6 N-terminal domain (112-164) and Med17 C-terminal domain (322-410 and 457-479) (Fig. S2). Distance constraints from MS/MS-coupled cross-linking analyses guided and validated new model build ( Fig. 1) (24) and supported revisions of previous secondary structure assignments in the Med11 N-terminal do- main ( Fig. S3) and in the Med17 C-terminal domain (Fig. S4). Selenomethionine anomalous difference maps were also used to ...
Context 2
... soaked with a 35-residue peptide containing five CTD heptapeptide repeats gave rise to difference electron density for the CTD (Fig. 2), whereas crystals soaked with a peptide con- taining only two repeats did not. Despite the moderate resolu- tion of the CTD difference maps (4.5 Å), clear structural fea- tures allow the sequence register and directionality of the CTD to be modeled with confidence. Bulky side-chain density attrib- utable to tyrosine residues was ...
Context 3
... soaked with a peptide con- taining only two repeats did not. Despite the moderate resolu- tion of the CTD difference maps (4.5 Å), clear structural fea- tures allow the sequence register and directionality of the CTD to be modeled with confidence. Bulky side-chain density attrib- utable to tyrosine residues was clearly visible in some places (Fig. 2C). A kinked central section of the difference density can be accounted for by a β-turn motif ( 2 SPTSPS 7 ), stabilized by an intramolecular hydrogen-bond network (25), seen in previous X- ray structures of CTD peptides (Fig. 2D). This placement of the β-turn motif was supported by the occurrence of bulky side-chain density for tyrosine ...
Context 4
... modeled with confidence. Bulky side-chain density attrib- utable to tyrosine residues was clearly visible in some places (Fig. 2C). A kinked central section of the difference density can be accounted for by a β-turn motif ( 2 SPTSPS 7 ), stabilized by an intramolecular hydrogen-bond network (25), seen in previous X- ray structures of CTD peptides (Fig. 2D). This placement of the β-turn motif was supported by the occurrence of bulky side-chain density for tyrosine residues located on either side (Fig. 2D). A CTD model was built by extending both ends of the β-turn motif, with sequence register constraints provided by bulky side-chain densities, and with all-trans peptide bond geometry ...
Context 5
... of the difference density can be accounted for by a β-turn motif ( 2 SPTSPS 7 ), stabilized by an intramolecular hydrogen-bond network (25), seen in previous X- ray structures of CTD peptides (Fig. 2D). This placement of the β-turn motif was supported by the occurrence of bulky side-chain density for tyrosine residues located on either side (Fig. 2D). A CTD model was built by extending both ends of the β-turn motif, with sequence register constraints provided by bulky side-chain densities, and with all-trans peptide bond geometry imposed, in keeping with structures of CTD peptides in complexes with RNA processing proteins (26,27). The entire CTD model comprises almost four ...
Similar publications
Homeodomain-interacting protein kinases (HIPKs) belong to the CMGC kinase family and are closely related to dual-specificity tyrosine phosphorylation-regulated kinases (DYRKs). HIPKs are regulators of various signaling pathways and involved in the pathology of cancer, chronic fibrosis, diabetes, and multiple neurodegenerative diseases. Here, we rep...



- [...]

TOX4 is one of the regulatory factors of PP1 phosphatases with poorly understood functions. Here we show that chromatin occupancy pattern of TOX4 resembles that of RNA polymerase II (Pol II), and its loss increases cellular level of C-terminal domain (CTD) phosphorylated Pol II but mainly decreases Pol II occupancy on promoters. In addition, elonga...
The Nrd1-Nab3-Sen1 (NNS) complex carries out the transcription termination of non-coding RNAs (ncRNAs) in yeast, although the detailed interactions among its subunits remain obscure. Here we have identified three sequence motifs in Sen1 that mediate direct interactions with the RNA polymerase II CTD interaction domain (CID) of Nrd1, determined the...
Significance
Rho GTPase and polymerase II (Pol II), two key molecules involved in cellular signaling and transcription in eukaryotic organisms, have been separately studied for more than 2 decades without evidence showing their functional linkage. We provide genetic and biochemical evidence linking these two molecules in an intracellular signaling...
Transitions between the different stages of the RNAPII transcription cycle involve the recruitment and exchange of factors,
including mRNA capping enzymes, elongation factors, splicing factors, 3′-end-processing complexes, and termination factors.
These transitions are coordinated by the dynamic phosphorylation of the C-terminal domain (CTD) of the...
Citations
... Next, we prepared the 1.37 MDa human Mediator complex (hMED) [29][30][31][32] to investigate co-recruitment between hMED and hCTD. Part of the hMed sample was fluorescently labeled with Alexa Flour 647. ...
Eukaryotic gene regulation and pre-mRNA transcription depend on the carboxy-terminal domain (CTD) of RNA polymerase (Pol) II. Due to its highly repetitive, intrinsically disordered sequence, the CTD enables clustering and phase separation of Pol II. The molecular interactions that drive CTD phase separation and Pol II clustering are unclear. Here, we show that multivalent interactions involving tyrosine impart temperature- and concentration-dependent self-coacervation of the CTD. NMR spectroscopy, molecular ensemble calculations and all-atom molecular dynamics simulations demonstrate the presence of diverse tyrosine-engaging interactions, including tyrosine-proline contacts, in condensed states of human CTD and other low-complexity proteins. We further show that the network of multivalent interactions involving tyrosine is responsible for the co-recruitment of the human Mediator complex and CTD during phase separation. Our work advances the understanding of the driving forces of CTD phase separation and thus provides the basis to better understand CTD-mediated Pol II clustering in eukaryotic gene transcription.
... Mediator from very early EM studies was found to comprise semidistinct "Head," "Middle," "Tail," and "Kinase" modules, an idea reinforced by the ability to obtain purifications of subsets of factors using biochemical and genetic manipulations. An earlier crystallography study soaking a 35-residue CTD peptide into crystals of the isolated S. cerevisiae Mediator "head" domain suggested a potential interface between the Mediator head and free CTD (14). Newer structures define a human Mediator-CTD binding region in the vicinity of the earlier proposed yeast site but do so in the context of the PIC and more complete human Mediator complexes (13,15). ...
... At the time of our last review of Mediator (Plaschka et al. 2016), several research groups had employed integrated structural biology methods to arrive at an atomic model for the Mediator head module (Lariviere et al. 2012;Robinson et al. 2012) and a topological model for the middle module (Lariviere et al. 2013;Robinson et al. 2015), which led to a composite model for core Mediator (cMed) (Lariviere et al. 2012(Lariviere et al. , 2013. The structures showed that the Mediator head module consists of eight submodulesthe shoulder, arm, spine, joint, moveable jaw, finger, tooth and nose ( Figure 1a) (Lariviere et al. 2012). ...
... Both studies confirmed the position of Mediator on the PIC, as reported previously (Plaschka et al. 2015) and were in agreement with respect to the location of the CDK-activating kinase (CAK) module of TFIIH between the shoulder and hook of the Mediator head and middle modules, respectively. Complemented by crosslinking data and previously determined crystal structures of the Mediator head module in complex with an Rpb1 CTD heptapeptide repeat (YSPTSPS), both studies derive the putative trajectory of the Rpb1 CTD towards the active site of the TFIIH kinase Kin28 (human CDK7) (Robinson et al. 2012). The study presenting the higher resolution structure also embossed the previously reported Mediator interactions with Pol II and the general transcription factor and PIC component, TFIIB (Plaschka et al. 2015). ...
Recent advances in cryo-electron microscopy have led to multiple structures of Mediator in complex with the RNA polymerase II (Pol II) transcription initiation machinery. As a result we now hold in hands near-complete structures of both yeast and human Mediator complexes and have a better understanding of their interactions with the Pol II pre-initiation complex (PIC). Herein, we provide a summary of recent achievements and discuss their implications for future studies of Mediator and its role in gene regulation.
... Des sous-structures ont parfois été obtenues séparément, comme la structure d'un module seul, ou à l'inverse, la structure du Médiateur a pu être obtenue aussi en interaction avec des composants du PIC. Les données sur la structure du Médiateur ont été obtenues par différentes méthodes : des analyses biochimiques couplées à une réticulation et une spectrométrie de masse, par cristallographie à rayons X à des résolutions d'environ 4.5 angströms, par microscopie cryoélectronique à des résolutions d'environ 4. 3, 9.7, 15, 18, 20 et 28 angströms (Cai et al., 2009;Imasaki et al., 2011;Plaschka et al., 2015;Robinson et al., 2015Robinson et al., , 2016Robinson et al., , 2012Sato et al., 2016;Tsai et al., 2013Tsai et al., , 2014Wang et al., 2014). Ces études structurales ont révélé que le complexe s'organise en 4 modules nommés la tête, le milieu, la queue, et le module kinase (figure 7), ce dernier s'associant de manière transitoire aux trois autres. ...
Dans les cellules eucaryotes, l'ADN est compacté sous forme de chromatine et tous les processus qui y sont liés y compris la transcription se déroulent dans ce contexte chromatinien. Le Médiateur de la régulation transcriptionnelle est un complexe multiprotéique essentiel et conservé qui intègre les signaux de régulation des facteurs de transcription et les transmet à la machinerie transcriptionnelle. Etant donné son rôle essentiel dans la transcription des gènes, des mutations qui l'affectent sont impliquées dans un large spectre de maladies comme le cancer. Bien que le Médiateur ait fait l'objet de nombreuses études, on ne sait pas comment il coordonne ses fonctions avec les autres corégulateurs transcriptionnels. Nous avons observé que plusieurs de ses sous-unités sont en contact avec le remodeleur de la chromatine RSC, très abondant dans la cellule et essentiel pour sa viabilité. L'importance de ce complexe est soulignée par la grande implication de son homologue humain PBAF dans les pathologies cancéreuses : 20 % des cancers portent une mutation dans au moins une sous-unité de cette famille de remodeleurs. Dans ce projet, nous avons étudié les mécanismes d'action du Médiateur en relation avec RSC dans la transcription. Nous avons caractérisé une interaction physique et fonctionnelle entre les deux complexes, en utilisant principalement la levure comme modèle d'étude. Ce projet représente un enjeu majeur dans le domaine de la transcription génique et nous permet de mieux comprendre ce processus cellulaire fondamental, impliqué dans l'établissement et le maintien de nombreuses maladies grave.
... Mediator toward the CAK module in the Mediator-PIC-TFIID structure ( Fig. 2B-C) [25]. This channel is formed by the knob domain (MED31 and MED14) and the shoulder and neck domains (MED6, MED8 and MED17), and a short CTD peptide had previously been mapped to the same location in a yeast Head module crystal [57]. The Mediator-PIC-TBP structure identified a smaller peptide similarly positioned in this channel, as well as another peptide fitting in the CDK7 active site and primed for Ser5 phosphorylation (Fig. 2B) [24]. ...
Mediator is a large modular protein assembly whose function as a coactivator of transcription is conserved in all eukaryotes. The Mediator complex can integrate and relay signals from gene‐specific activators bound at enhancers to activate the general transcription machinery located at promoters. It has thus been described as a bridge between these elements during initiation of transcription. Here, we review recent studies on Mediator relating to its structure, gene specificity and general requirement, roles in chromatin architecture as well as novel concepts involving phase separation and transcriptional bursting. We revisit the mechanism of action of Mediator and ultimately put forward models for its mode of action in gene activation.
... Interface D is formed by contacts between the N-terminal region of the MED6 shoulder domain and two CDK7 surface regions (around residues 103-110 and 259-266) that differ in other kinases of the CDK family 33 . Interface E is formed between the hook domain of the middle module-including the MED14 N-terminal region that comprises helices α1 and α2-and the CDK7 N-terminal region (residues [10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26]. This N-terminal region of CDK7 flanks the ATP-binding site and active centre. ...
Mediator is a conserved coactivator that enables regulated transcription initiation at eukaryotic genes1–3. Mediator is recruited by transcriptional activators and binds the pre-initiation complex (PIC) to stimulate RNA polymerase II (Pol II) phosphorylation and promoter escape1–6. Here we prepare a recombinant human Mediator, reconstitute a 50-subunit Mediator-PIC complex, and determine the structure of the complex by cryo-EM. Mediator uses its head module to contact the Pol II stalk and the general transcription factors TFIIB and TFIIE, resembling the Mediator-PIC interactions in the corresponding yeast complex7–9. The metazoan subunits MED27-MED30 associate with exposed regions in MED14 and MED17 to form the proximal part of the Mediator tail module that binds activators. Mediator positions the flexibly linked CDK-activating kinase (CAK) of the general transcription factor TFIIH near the linker to the C-terminal repeat domain (CTD) of Pol II. The Mediator shoulder domain holds the CAK subunit CDK7, whereas the hook domain contacts a CDK7 element that flanks the kinase active site. The shoulder and hook reside in the Mediator head and middle modules, respectively, which can move relative to each other and may induce an active conformation of the CDK7 kinase to allosterically stimulate CTD phosphorylation.
... Although the Tyr 1 , Pro 3 , and Pro 6 residues fit well into the cryo-EM map, the repetitive CTD is almost symmetric (Y 1 S 2 P 3 T 4 S 5 P 6 S 7 Y 1 S 2 P 3 T 4 S 5 P 6 S 7 Y 1 ) and the CTD direction could not be unambiguously determined solely from the cryo-EM map. The directions of the two CTD segments were proposed according to their topological placement relative to Pol II and the structure of CTD in complex with the Head of yMed (70). ...
... S22G). The CTD-binding pattern differs from that observed in the crystal structure of CTD-Head of yMed (70), possibly because of the lack of stabilization by the Knob in the crystal structure ( fig. S22, E and F). ...
... The models of CTD-Mediator and CAK were built according to the locally refined maps around CTD-Mediator at 3.7 Å resolution and CAK at 4.5 Å resolution in COOT (90). The direction of the two peptides were proposed according to the previously determined crystal structure of CTD-Head (70) and their topological positions relative to Pol II. All the structures were refined in real space using Phenix (91) with secondary structure and geometry restraints. ...
A complete PIC-Mediator structure
As a critical transcription coactivator, the multisubunit Mediator complex binds RNA polymerase II (Pol II), facilitates preinitiation complex (PIC) assembly, and stimulates transcription and phosphorylation of the Pol II C-terminal domain (CTD). However, how these critical transcriptional events are coordinated by Mediator is not fully understood. Chen et al. determined the structures of human Mediator and Mediator-bound PIC in distinct conformational states, the latter of which represents a complete PIC-Mediator complex assembled on the 14-subunit transcription factor IID (TFIID). The structures show that Mediator undergoes reorganization during PIC-Mediator assembly, sandwiches and facilitates phosphorylation of Pol II CTD, and works with TFIID to organize TFIIH in PIC for transcription initiation.
Science , abg0635, this issue p. eabg0635
... One such protein is Mediator, which is in fact a large set of protein factors that interact with RNA pol II and DNA regulator sequences distant from the transcribed gene to strongly enhance or repress transcription [29]. When bound to Mediator, the CTD of RNA pol II adopts two PPII helices connected by a turn [30]. Work over the last 15 years has shown that the Mediator complex is essential for forming a phase-separated 'superenhancer' condensate, which dramatically increases transcription rates and plays important physiological roles as well as in cancer [31]. ...
Biomolecular condensates are microdroplets that form inside cells and serve to selectively concentrate proteins, RNAs and other molecules for a variety of physiological functions, but can contribute to cancer, neurodegenerative diseases and viral infections. The formation of these condensates is driven by weak, transient interactions between molecules. These weak associations can operate at the level of whole protein domains, elements of secondary structure or even moieties composed of just a few atoms. Different types of condensates do not generally combine to form larger microdroplets, suggesting that each uses a distinct class of attractive interactions. Here, we address whether polyproline II (PPII) helices mediate condensate formation. By combining with PPII‐binding elements such as GYF, WW, profilin, SH3 or OCRE domains, PPII helices help form lipid rafts, nuclear speckles, P‐body‐like neuronal granules, enhancer complexes and other condensates. The number of PPII helical tracts or tandem PPII‐binding domains can strongly influence condensate stability. Many PPII helices have a low content of proline residues, which hinders their identification. Recently, we characterized the NMR spectral properties of a Gly‐rich, Pro‐poor protein composed of six PPII helices. Based on those results, we predicted that many Gly‐rich segments may form PPII helices and interact with PPII‐binding domains. This prediction is being tested and could join the palette of verified interactions contributing to biomolecular condensate formation. Polyproline II helices and the domains that bind them are common in biology. Often connected in tandem, their associations can drive the formation of biomacromolecular condensates. Glycine‐rich polyproline II helices can combine to form polyproline II helical bundles and might then associate with drive phase transitions.
... The active site of TFIIK lies on the inner wall of the tunnel, so that CTD phosphorylation may be processive as it threads through the tunnel (5,19). The ~25-residue segment of CTD bound to the Mediator head in the CTD channel (19,43) is oriented for the delivery to the active site of TFIIK. When a sevenresidue CTD (SPTSPSY) is modeled on TFIIK (Fig. 6, right) N-terminal end is ~37 Å away from the C-terminal end of the CTD segment bound to the Mediator head and may be connected by simply extending ~13 residues. ...
During transcription initiation, the general transcription factor TFIIH marks RNA polymerase II by phosphorylating Ser5 of the carboxyl-terminal domain (CTD) of Rpb1, which is followed by extensive modifications coupled to transcription elongation, mRNA processing, and histone dynamics. We have determined a 3.5-Å resolution cryo–electron microscopy (cryo-EM) structure of the TFIIH kinase module (TFIIK in yeast), which is composed of Kin28, Ccl1, and Tfb3, yeast homologs of CDK7, cyclin H, and MAT1, respectively. The carboxyl-terminal region of Tfb3 was lying at the edge of catalytic cleft of Kin28, where a conserved Tfb3 helix served to stabilize the activation loop in its active conformation. By combining the structure of TFIIK with the previous cryo-EM structure of the preinitiation complex, we extend the previously proposed model of the CTD path to the active site of TFIIK.
... The existing high-resolution models of yMed-PIC highlight differences in how MedHead interacts with Pol II, suggesting that the interface between Mediator and Pol II is not rigid (6,7). MedHead is capable of binding the CTD, shown in a co-crystal structure (17). Based on the location of the CTD modeled into the full yeast Mediator complex, the CTD also likely serves to stabilize the interface between MedHead and MedMiddle (8,9). ...
... S. cerevisiae MedHead (scMedHead) was co-crystallized with a short peptide of the RPB1 CTD, which shows slightly more than three full repeats engaged with scMedHead at the shoulder and neck domains (17). We observe additional electron density in this same location and used the S. cerevisiae structure to build a model for this portion of the CTD that we will refer to as MED CTD (Fig. 5D-E). ...
Mediating transcription
The Mediator complex is recruited by transcription factors to all protein-coding genes in eukaryotes and helps to assemble the machinery necessary to transcribe the gene. Abdella et al. present the cryo–electron microscopy structure of the human Mediator-bound preinitiation complex (Med-PIC). The structure shows how Mediator positions the long, flexible C-terminal domain of RNA polymerase II to be phosphorylated by the kinase CDK7, a crucial step for further processing of the RNA into a mature RNA. Most sites where transcription factors bind to Mediator are flexibly tethered to the complex, allowing the large Med-PIC to assemble at any gene.
Science , this issue p. 52