Fig 3 - available via license: Creative Commons Attribution 4.0 International
Content may be subject to copyright.
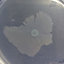
Parallel evolution of endpoint populations was observed from Trials 1 and 2. Venn diagrams represent the number of (A) all mutations, (B) nsSNPs, or (C) genes that were observed in the individual trials (Trial 1, green; Trial 2, blue).
Source publication
The evolution of hypermutators in response to antibiotic treatment in both clinical and laboratory settings provides a unique context for the study of adaptive evolution. With increased mutation rates, the number of hitchhiker mutations within an evolving hypermutator population is remarkably high and presents substantial challenges in determining...
Similar publications
Adaptive laboratory evolution is an important tool to evolve organisms to increased tolerance towards different physical and chemical stress. It is applied to study the evolution of antibiotic resistance as well as genetic mechanisms underlying improvements in production strains. Adaptive evolution experiments can be automated in a high-throughput...
Citations
... Hypermutation (i.e., an observed increase in mutation rates) has been commonly observed in microorganisms when placed under great selective pressure, especially in pathogens (Swings et al., 2017;Wiser et al., 2013). A much higher prevalence of hypermutators has been found to exist in natural bacterial populations (Gross & Siegel, 1981;Hall & Henderson-Begg, 2006) such as E. coli (LeClerc et al., 1996;Matic et al., 1997;Denamur et al., 2006), Pseudomonas aeruginosa (Marvig et al., 2015;Oliver, 2015), Salmonella (LeClerc et al., 1996), Staphylococcus aureus (Iguchi et al., 2016), A. baumannii (Hammerstrom et al., 2015), and others (see for more Swings et al., 2017). The high mutation rate of HIV has also allowed for its continual evasion of the immune system (Rambaut et al., 2004). ...
Change is the fundamental idea of evolution. Explaining the extraordinary biological change we see written in the history of genomes and fossil beds is the primary occupation of the evolutionary biologist. Yet it is a surprising fact that for the majority of evolutionary research, we have rarely studied how evolution typically unfolds in nature, in changing ecological environments, over space and time. While ecology played a major role in the eventual acceptance of the population genetic viewpoint of evolution in the synthetic era (circa 1918-1956), it held a lesser role in the development of evolutionary theory until the 1980s, when we began to systematically study the evolutionary dynamics of natural populations in space and time. As a result, early evolutionary theory was initially constructed in an abstract vacuum that was unrepresentative of evolution in nature. The subtle synthesis between ecology with evolutionary biology (eco-evo synthesis) over the past 40 years has progressed our knowledge of natural selection dynamics as they are found in nature, thus revealing how natural selection varies in strength, direction, form, and, more surprisingly, level of biological organization. Natural selection can no longer be reduced to lower levels of biological organization (i.e., individuals, selfish genes) over shorter timescales but should be expanded to include adaptation at higher levels and over longer timescales. Long-term and/or emergent evolutionary phenomena, such as multilevel selection or evolvability, have thus become tenable concepts within an evolutionary biology that embraces ecology and spatiotemporal change.
Evolutionary biology is currently suspended at an intermediate stage of scientific progress that calls for the organization of all the recent knowledge revealed by the eco-evo synthesis into a coherent and unified theoretical framework. This is where philosophers of biology can be of particular use, acting as a bridge between the subdisciplines of biology and inventing new theoretical strategies to organize and accommodate the recent knowledge. Philosophers have recommended transitioning away from outdated philosophies that were originally derived from physics within the philosophical zeitgeist of logical positivism (i.e., monism, reductionism, and monocausation) and toward a distinct philosophy of biology that can capture the natural complexity of multifaceted biological systems within diverse ecosystems—one that embraces the emerging philosophies of pluralism, emergence, and multicausality.
Therefore, I see recent advances in ecology, evolutionary biology, and the philosophy of biology as laying the groundwork for another major biological synthesis, what I refer to as the Second Synthesis because, in many respects, it is analogous to the aims and outcomes of the first major biological synthesis (but is notably distinct from the inorganic and contrived progressive movement known as the extended evolutionary synthesis). With the general development of a distinctive philosophy of science, biology has rightfully emerged as an autonomous science. Thus, while the first synthesis legitimized biology, the Second Synthesis autonomized biology and afforded biology its own philosophy, allowing biology to finally realize its full scientific potential.
... The same study also showed that hypermutators have a competitive advantage over normomutators in the presence of this phage, but not in the absence of it. Other studies also noted a higher occurrence of hypermutators in experimental populations adapting to antibiotic stress (Hammerstrom et al., 2015;Mao et al., 1997). Finally, Swings et al. (2017) showed that the occurrence of hypermutators allows Escherichia coli populations to adapt to higher concentrations of ethanol compared to populations where no hypermutators appeared. ...
... The observed increase in hypermutator emergence due to osmotic stress is in line with other studies where maladaptation due to an environmental stressor increased the rate of hypermutator emergence (Pal et al., 2007;Swings et al., 2017). The finding that, at least under our experimental conditions, the emergence of hypermutators is not affected by exposure to gentamicin furthermore nuances earlier reports on the evolution of increased mutation rates under antibiotic stress (Hammerstrom et al., 2015;Ibacache-Quiroga et al., 2018). Although in our experiment hypermutators frequently evolved when exposed to osmotic stress, hypermutator populations did not show a detectable increase in the rate or degree of adaptation to osmotic stress. ...
Genotypes exhibiting an increased mutation rate, called hypermutators, can propagate in microbial populations because they can have an advantage due to the higher supply of beneficial mutations needed for adaptation. Although this is a frequently observed phenomenon in natural and laboratory populations, little is known about the influence of parameters such as the degree of maladaptation, stress intensity, and the genetic architecture for adaptation on the emergence of hypermutators. To address this knowledge gap, we measured the emergence of hypermutators over ~1,000 generations in experimental Escherichia coli populations exposed to different levels of osmotic or antibiotic stress. Our stress types were chosen based on the assumption that the genetic architecture for adaptation differs between them. Indeed, we show that the size of the genetic basis for adaptation is larger for osmotic stress compared to antibiotic stress. During our experiment, we observed an increased emergence of hypermutators in populations exposed to osmotic stress but not in those exposed to antibiotic stress, indicating that hypermutator emergence rates are stress type dependent. These results support our hypothesis that hypermutator emergence is linked to the size of the genetic basis for adaptation. In addition, we identified other parameters that covaried with stress type (stress level and IS transposition rates) that might have contributed to an increased hypermutator provision and selection. Our results provide a first comparison of hypermutator emergence rates under varying stress conditions and point towards complex interactions of multiple stress-related factors on the evolution of mutation rates.
... adeB expression was significantly increased in all 13 tigecycline-insensitive strains, and detection of adeR and adeS genes in these strains revealed by comparison that they had 2-7 nonsynonymous mutations. The adeS gene was more prone to mutations than the adeR gene, a phenomenon in agreement with the studies of Montana S et al. [40] and Hammerstrom Troy G et al. [41], suggesting that adeS has a higher degree of genetic variation and plays a more active role in the regulation of adeABC overexpression. Four nonsynonymous point mutations (Val120Ile, Ala136Val, Gly232Ala, Asp20Asn) and one insertional mutation (Insert32C) were found in adeR gene, and 12 nonsynonymous point mutations (Ala94Val, Ala97Thr, Leu105Phe, Leu172Pro, Gly186Val Arg195Gln, Gln203Leu, Asn268His, Tyr303Phe, Lys315Asn, Gly319Ser, Val348Ile) and one insertional mutation (ISAba1 inserted at the end of adeS) were found in adeS gene. ...
Background:
The isolation of tigecycline-resistant Acinetobacter baumannii in recent years has brought great difficulties to clinical prevention and treatment.
Purpose:
To explore the effect of efflux pump system and other resistance related gene mutations on tigecycline resistance in Acinetobacter baumannii.
Methods:
Fluorescence quantitative PCR was used to detect the expression levels of major efflux pump genes (adeB, adeJ, and adeG) in extensive drug-resistant Acinetobacter baumannii. The minimum inhibitory concentration (MIC) of tigecycline was detected by the broth microdilution testing and efflux pump inhibition experiment to assess the role of efflux pump in tigecycline resistance of Acinetobacter baumannii. Efflux pump regulatory genes (adeR and adeS) and tigecycline resistance related genes (rpsJ, trm, and plsC) were amplified by PCR and sequenced. By sequence alignment, tigecycline sensitive and tigecycline-insensitive Acinetobacter baumannii were compared with standard strains to analyze the presence of mutations in these genes.
Results:
The relative expression of adeB in the tigecycline-insensitive Acinetobacter baumannii was significantly higher than that in the tigecycline sensitive Acinetobacter baumannii (114.70 (89.53-157.43) vs 86.12 (27.23-129.34), P = 0.025). When efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was added, the percentage of tigecycline-insensitive Acinetobacter baumannii with tigecycline MIC decreased was significantly higher than that of tigecycline-sensitive Acinetobacter baumannii (10/13 (76.9%) vs 26/59 (44.1%)), P = 0.032); the relative expression of adeB in the MIC decreased group was significantly higher than that in the MIC unchanged group (110.29 (63.62-147.15) vs 50.06 (26.10-122.59), P = 0.02); The relative expression levels of efflux pumps adeG and adeJ did not increase significantly, and there was no significant difference between these groups. One adeR point mutation (Gly232Ala) and eight adeS point mutations (Ala97Thr, Leu105Phe, Leu172Pro, Arg195Gln, Gln203Leu, Tyr303Phe, Lys315Asn, Gly319Ser) were newly detected. Consistent mutations in trm and plsC genes were detected in both tigecycline-insensitive and tigecycline-sensitive Acinetobacter baumannii, but no mutation in rpsJ gene was detected in them.
Conclusion:
Tigecycline-insensitive Acinetobacter baumannii efflux pump adeABC overexpression was an important mechanism for tigecycline resistance, and the mutations of efflux pump regulator genes (adeR and adeS) are responsible for adeABC overexpression. The effect of trm, plsC, and rpsJ gene mutations on the development of tigecycline resistance in Acinetobacter baumannii remains controversial.
... While technically more demanding, an advantage of these systems is that the biofilm does not have to be dispersed in between different treatment cycles. Examples include acrylic flow cells with a glass surface 80 , Sartorius bioreactors [81][82][83] and various microfluidic devices [84][85][86] . ...
... Finally, the treatment regime can have an influence on the evolutionary trajectory that is followed. The concentration of the antimicrobial agent can be kept constant during the course of the evolution experiment 70,75 or bacteria can be exposed to gradually increasing antimicrobial concentrations 82,85 , and exposure can be continuous 69,74,75,82,85 or intermittent 70,77,103,104 . Regrowth of the biofilm after each treatment cycle ensures that biofilms with similar cell densities are studied throughout the experiment, and the regrowth phase can mimic the decrease of antibiotic concentration in between two treatments. ...
... Finally, the treatment regime can have an influence on the evolutionary trajectory that is followed. The concentration of the antimicrobial agent can be kept constant during the course of the evolution experiment 70,75 or bacteria can be exposed to gradually increasing antimicrobial concentrations 82,85 , and exposure can be continuous 69,74,75,82,85 or intermittent 70,77,103,104 . Regrowth of the biofilm after each treatment cycle ensures that biofilms with similar cell densities are studied throughout the experiment, and the regrowth phase can mimic the decrease of antibiotic concentration in between two treatments. ...
Experimental evolution experiments in which bacterial populations are repeatedly exposed to an antimicrobial treatment, and examination of the genotype and phenotype of the resulting evolved bacteria, can help shed light on mechanisms behind reduced susceptibility. In this review we present an overview of why it is important to include biofilms in experimental evolution, which approaches are available to study experimental evolution in biofilms and what experimental evolution has taught us about tolerance and resistance in biofilms. Finally, we present an emerging consensus view on biofilm antimicrobial susceptibility supported by data obtained during experimental evolution studies.
... In future, to constructively speculate the evolutionary trajectories accessible by hypermutators, the intermediate isolates should be genome-sequenced. Then, the genotype/allele frequency against generation is plotted to present the clonal interference of the inducible hypermutator in the vessel (which mimics CF lungs) (296,355). The dominating genotypes in vitro will then be analysed in reference to the clinical isolates to understand their chance of occurrence and prevalence. ...
Hypermutation occurs when the mismatch repair (MMR) system becomes defective due to e.g., mutation. In Pseudomonas aeruginosa (PA), the mutS gene is commonly mutated in hypermutator strains. MutS mutants display an elevated mutation rate (>100-fold, compared with the wild-type (WT)) and are often associated with the acquisition of antibiotic resistance in clinical PA isolates. In the current work, I construct a strain (PAYY01) of PA in which the expression of mutS (and therefore, the mutation frequency of the strain) can be modulated through the addition of an inert chemical inducer, rhamnose. In PAYY01, the mutation rate can be set to any desired value – either higher or lower than the WT, by adding an appropriate concentration of rhamnose. Indeed, and to my knowledge, this is the first time that a state of controlled hypomutability in PA has been reported. I therefore have a tool that allows me to either “step on the evolutionary accelerator” or “slam on the evolutionary brakes”, and this has opened up a wealth of research avenues to explore. For example, I used whole genome sequencing to investigate the impact of hypermutation during growth of PAYY01 in artificial sputum medium (ASM) and Luria Broth (LB). Very low levels of mutation were observed when mutS expression was “on”, whereas very high levels of mutation were observed when mutS expression was “off”. I also examined how induced hypermutability affects the interaction of PA with Staphylococcus aureus (SA) during co-culture. Somewhat surprisingly, the hypermutator was less invasive than the WT in batch culture during both inter- and intra-species competitions. By contrast, the hypermutator demonstrated a better ability to survive antibiotic (colistin) challenge than the WT. Overall, my data indicate that controlled mutability is an important tool in our efforts to better understand the evolution of drug resistance, inter-species interactions, and pathoadaptation. In addition, I also explore the architecture of the gene cluster surrounding mutS, and find that mutS forms an operon with the adjacent fdxA gene. This, in turn, stimulated an investigation of the possible participation of FdxA in mismatch detection/repair.
... These mutations were predominantly observed within the histidine kinase, adenylyl cyclase, methyl-accepting protein, and phosphatase (HAMP) and DHp domain (20). Of those, many substitutions were found adjacent or in close proximity to the autophosphorylation site H149, e.g., Q141R, R152K, T153A, T153M, and D167N, indicating a mutational hot spot (17)(18)(19)27). Investigation of AdeS T156M was based on a clinical isolate pair described in a previous study, where the two isolates were identical except for the T156M amino acid substitution in AdeS and susceptibility to tigecycline (17). ...
The active efflux of antimicrobials by bacteria can lead to antimicrobial resistance and persistence and can affect multiple different classes of antimicrobials. Efflux pumps are tightly regulated, and their overexpression can be mediated by changes in their regulators.
... These pumps also play key roles in bacterial virulence and physiology (12). The development of resistance in A. baumannii is driven largely by its existence in the hospital environment, where the use of a diverse array of antimicrobial compounds has been associated with constitutive upregulation of genes encoding RND efflux systems, particularly adeABC and adeIJK, via regulatory mutations (11,14,15). Specifically, selective pressure exerted by antimicrobials selects for direct mutations within the efflux systems or their corresponding regulatory components, including adeRS (adeABC) and adeN (adeIJK) (11,14,16,17). ...
The global distribution of multidrug resistance in A. baumannii has necessitated seeking not only alternative therapeutic approaches but also the means to limit the development of resistance in clinical settings. Highly abundant host bioactive compounds, such as polyunsaturated fatty acids, are readily acquired by A. baumannii during infection and have been illustrated to impact the bacterium’s membrane composition and antibiotic resistance.
... In addition, tigecycline evades tetracycline resistance mechanisms because its binding orientation is different from that of tetracyclines (6). Although tigecycline is regarded as a last-line antibiotic against infections caused by MDR or extensively drug-resistant (XDR) bacterial pathogens, tigecycline resistance has been reported worldwide (7). A. baumannii acquires tigecycline resistance by overexpression of efflux pumps, especially AdeABC, and modification of tigecycline-binding sites in ribosomes by rpsJ mutation (8). ...
... Many studies have suggested that resistance-nodulation-division (RND) efflux pumps, including AdeABC, are associated with tigecycline resistance (8,17). We confirmed the role of RND efflux pumps in tigecycline-resistant populations of heteroresistant A. baumannii isolates. ...
... The survival rate of the tigecycline-resistant subpopulation was measured after exposure or nonexposure to subinhibitory concentrations of tigecycline according to the method of a previous study (25), with some modifications. Overnight cultures of tigecycline-heteroresistant, -susceptible, and -intermediate, A. baumannii isolates grown in MH II broth were inoculated at a 1:100 ratio into fresh medium and treated with tigecycline at 0, 0.25, 0.5, 1, 2,4,8, and 16 mg/liter. After incubation for 24 h at 37°C with shaking at 185 rpm, each bacterial culture was inoculated onto MH II broth with 8 mg/liter of tigecycline to identify the influence of treatments with diverse concentrations on the selection of resistant subpopulations among the whole ...
Tigecycline is regarded as a last-resort treatment for multidrug-resistant Acinetobacter baumannii. However, tigecycline resistance in A. baumannii has increased worldwide. In this study, we investigated tigecycline heteroresistance in A. baumannii isolates from South Korea. Antibiotic susceptibility testing was performed on 323 nonduplicated A. baumannii isolates. Among 260 and 37 tigecycline-susceptible and -intermediate-resistant A. baumannii isolates, 146 (56.2%) and 22 (59.5%) isolates were identified as heteroresistant to tigecycline through a disk diffusion assay and population analysis profiling. For selected isolates, an in vitro time-kill assay was performed, and survival rates were measured after preincubation with diverse concentrations of tigecycline. Heteroresistant isolates showed regrowth after 12 h of 2_ MIC of tigecycline treatment, and resistant subpopulations were selected by preexposure to tigecycline. Furthermore, genetic alterations in adeABC, adeRS, and rpsJ were assessed, and the relative mRNA expression levels of adeB and adeS were compared. The tigecycline resistance in subpopulations might be due to the insertion of ISAba1 in adeS, leading to the overexpression of the AdeABC efflux pump. However, the tigecycline resistance of subpopulations was not stable during serial passages in antibiotic-free medium. The reversion of tigecycline susceptibility by antibiotic-free passages might occur by additional insertions of ISAba10 in adeR and nucleotide alterations in adeS in some mutants. Tigecycline heteroresistance is prevalent in A. baumannii isolates, which results in treatment failure. Tigecycline resistance is mainly due to the overexpression of the AdeABC efflux pump, which is associated with genetic mutations, but this resistance could be reversed into susceptibility by additional mutations in antibiotic-free environments.
... Experimental evolution research has demonstrated the possibility that the emergence of a mutator might occur under antibiotic exposure by the reversible insertion of a mobile element to inactivate mutS, resulting in several mutations independently able to increase resistance (at various levels) to a challenging antibiotic in the population, thus providing an "efficient survey" of potentially successful evolutionary pathways (109). It has been shown, both in the case of mutation-based resistance (110) and in the evolution of resistant genes carried by MGEs (111), that antibiotic-resistant organisms frequently have increased mutation rates, which suggests the evolutionary consequences of hypermutation. Does the fact that organisms with mutator alleles can hitchhike with antibiotic resistance phenotypes indicate that the rise in AMR might increase the evolvability of bacterial populations in general? ...
... In contrast, in large populations with higher mutation rates, the most successful strategy follows slower trajectories, such as the D240G mutation, a "survival of the flattest" dynamics (265,269), because under these conditions the fittest organisms are those showing the greatest robustness against the deleterious mutations (270). According to clinical evidence, the survival of the flattest in AMR is generally the most successful strategy, because the antibiotic bottlenecks select microorganisms with high mutation rates (110). ...
Evolution is the hallmark of life. Descriptions of the evolution of microorganisms have provided a wealth of information, but knowledge regarding "what happened" has precluded a deeper understanding of "how" evolution has proceeded, as in the case of antimicrobial resistance. The difficulty in answering the "how" question lies in the multihierarchical dimensions of evolutionary processes, nested in complex networks, encompassing all units of selection, from genes to communities and ecosystems. At the simplest ontological level (as resistance genes), evolution proceeds by random (mutation and drift) and directional (natural selection) processes; however, sequential pathways of adaptive variation can occasionally be observed, and under fixed circumstances (particular fitness landscapes), evolution is predictable. At the highest level (such as that of plasmids, clones, species, microbiotas), the systems' degrees of freedom increase dramatically, related to the variable dispersal, fragmentation, relatedness, or coalescence of bacterial populations, depending on heterogeneous and changing niches and selective gradients in complex environments. Evolutionary trajectories of antibiotic resistance find their way in these changing landscapes subjected to random variations, becoming highly entropic and therefore unpredictable. However, experimental, phylogenetic, and ecogenetic analyses reveal preferential frequented paths (highways) where antibiotic resistance flows and propagates, allowing some understanding of evolutionary dynamics, modeling and designing interventions. Studies on antibiotic resistance have an applied aspect in improving individual health, One Health, and Global Health, as well as an academic value for understanding evolution. Most importantly, they have a heuristic significance as a model to reduce the negative influence of anthropogenic effects on the environment.
... Generally, the mutation rate in bacteria with functional DNA repair enzymes is about 0.001-0.003 mutation per genome per cell division, whereas there are also hypermutator bacteria, with orders of magnitude higher mutation rates than those of DNA-repair functional strains (Drake 1991;Oliver et al. 2000;Chopra et al. 2003;Foster 2007;Lee et al. 2012;Hammerstrom et al. 2015;Long, Sung, et al. 2015). The mutation rate of cells with MMR deficiency is greatly elevated in both prokaryotes and eukaryotes, especially in terms of transitions (Kunkel and Erie 2005). ...
Microbial strains with high genomic stability are particularly sought after for testing the quality of commercial microbiological products, such as biological media and antibiotics. Yet, using mutation-accumulation experiments and de novo assembled complete genomes based on Nanopore long-read sequencing, we find that the widely-used quality-control strain S. putrefaciens ATCC-8071, also a facultative pathogen, is a hypermutator, with a base-pair substitution (BPS) mutation rate of 2.42 × 10−8 per nucleotide site per cell division, ∼146-fold greater than that of the wild-type strain CGMCC-1.6515. Using complementation experiments, we confirm that mutL dysfunction, which was a recent evolutionary event, is the cause for the high mutation rate of ATCC-8071. Further analyses also give insight into possible relationships between mutation and genome evolution in this important bacterium. This discovery of a well-known strain being a hypermutator necessitates screening the mutation rate of bacterial strains before any quality control or experiments.