Figure - available via license: CC BY
Content may be subject to copyright.
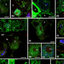
Immunogold localization of snRNP-indicating molecules in nuclear bodies of microspore and vegetative cell of young pollen grain (N-nucleus, No - nucleolus, VN-vegetative nucleus). Bars-0.5 μm. Figure 4. Immunogold labelling of TMG snRNA in the vegetative nucleus of young pollen grain is shown. Strong labelling can be observed in the nuclear body (arrowhead) localized in close proximity to the nucleolus. Figure 4a. Ultrastructural Sm protein localization in the vegetative nucleus of young pollen grain is shown. Numerous gold particles are concentrated over the nuclear body (arrowhead). Figure 4b. In situ hybridization to U2 snRNA is shown. The presence of U2 snRNA over the nuclear body can be seen in the microspore nucleus (arrowhead). Figure 4c. DNA labelling using TdT technique in polarized microspore nucleus (N-nucleus) is shown. No labelling can be seen in the nuclear body (arrowhead). Figure 4d. Control reaction of in situ hybridization performed using U2 snRNA sense probe (N-nucleus, No-nucleolus) is shown. No gold particles can be seen over the nucleoplasm and nuclear body (arrowhead) localized in close proximity to the nucleolus.
Source publication
The aim of the present work was the characterization of nuclear bodies in the microspore and developing pollen cells of Hyacinthus orientalis L.. The combination of Ag-NOR, immunofluorescence and immunogold techniques was used in this study. The obtained results showed the presence of highly agyrophylic extranucleolar bodies in microspore and devel...
Citations
... Larch microsporocytes likely accumulate and store snRNAs during late diplotene that will later be transmitted to the dyad cells during cell division (the half-life of snRNAs is long, often exceeding the cell life cycle) [39]. The large snRNA quantities that are synthesised and stored during the diplotene stage likely ensure that the cells contain the splicing machinery required during the development of a very transcriptionally active microspore [40][41][42][43][44]. Additionally, some snRNAs present in the larch microspore nucleus after cell division [45] are synthesised during the diplotene stage, and a high nuclear content of splicing factors has been observed in the early stages of H. orientalis microspores and Brassica napus [46][47][48]. The snRNA accumulation and transmission to dyads cells has also been observed in Danio rerio embryos, in which snRNAs are present in CBs at the 4-cell stage, which occurs 1 hour after fertilisation, and before zygotic genome activation at the 512-cell stage, which occurs approximately 2.75 hours after fertilisation [49]. ...
Manuscript provides insights into the biology of long-lived plants, different from Arabidopsis, tomato or grass species that are widely studied. In the European larch the diplotene stage lasts approximately 5 months and it is possible to divide it into several substages and to observe each of them in details. The diplotene stage is a period of intensive microsporocyte growth associated with the synthesis and accumulation of different RNA and proteins. Larch microsporocytes display changes in chromatin morphology during thus stage, alternating between 4 short stages of chromatin condensation (contraction) and 5 longer diffusion (relaxation) stages. The occurrence of a diplotene diffusion stage has been observed in many plant species. Interestingly, they has also been observed during spermiogenesis and oogenesis in animals.
The aim of this study was to examine whether chromatin relaxation during the diplotene is accompanied by the synthesis and maturation of mRNA.
The results reveal a correlation between the diffusion and chromatin decondensation, transcriptional activity. We also found decreasing amount of poly(A) mRNA synthesis in the consecutive diffusion stages. During the early diffusion stages, mRNA is intensively synthesized. In the nuclei large amounts of RNA polymerase II, and high levels of snRNPs was observed. In the late diffusion stages, the synthesized mRNA is not directly subjected to translation but it is stored in the nucleus, and later transported to the cytoplasm and translated. In the last diffusion stage, the level of poly(A) RNA is low, but that of splicing factors is still high.
It appears that the mRNA synthesized in early stages is used during the diplotene stage and is not transmitted to dyad and tetrads. In contrast, splicing factors accumulate and are most likely transmitted to the dyad and tetrads, where they are used after the resumption of intense transcription. Similar meiotic process were observed during oogenesis in animals. This indicates the existence of an evolutionarily conserved mechanism of chromatin-based regulation of gene expression during meiotic prophase I.
... After maceration, flowers were impregnated by 50% AgNO3 in water, 55 °C, 20 min; then incubated in 1:1 mixture of 2% gelatin in 1% formic acid and 50% AgNO3 for 20 min in dark. The plant material was then washed with distilled water for 10 min and then treated with 5% sodium thiosulfate for 10 min (Zienkiewicz & Bednarska 2009;modified). Individual anthers of sequential developmental series were prepared on glass slide, covered with water, squashed and observed under the microscope (Nicon Eclipse TE2000-E). The area of nucleolus was measured by NIS-Elements AR 3.0 software tools (Nikon Instruments, Tokyo, Japan) by hand selection at high magnification to avoid boundary errors. ...
... Initially, we used modified AgNOR staining to visualize nuclear bodies and Cajal bodies in particular (Zienkiewicz & Bednarska 2009) since REN1 protein co-localized there as well. Unfortunately, we were not able to observe these structures in Arabidopsis pollen probably due to smaller size of pollen grains and their rougher surface than in Hyacinthus for which this protocol was originally developed. ...
Heat shock transcription factors (Hsfs) are involved in multiple aspects of stress response and plant growth. However, their role during male gametophyte development is largely unknown, although the generative phase is the most sensitive and critical period in the plant life cycle. Based on a wide screen of T-DNA mutant lines, we identified the atren1 mutation (restricted to nucleolus1) in early male gametophytic gene At1g77570 which has the closest homology to HSFA5 gene, the member of a heat shock transcription factor (HSF) gene family. The mutation causes multiple defects in male gametophyte development in both structure and function. Since the mutation disrupts an early acting (AtREN1) gene, these pollen phenotype abnormalities appear from bicellular pollen stage to pollen maturation. Moreover, the consequent progamic phase is compromised as well as documented by pollen germination defects and limited transmission via male gametophyte. In addition, atren1/- plants are defective in heat stress (HS) response and produce notably higher proportion of aberrant pollen grains. AtREN1 protein is targeted specifically to the nucleolus that, together with the increased size of the nucleolus in atren1 pollen, suggests that it is likely to be involved in ribosomal RNA biogenesis or other nucleolar functions.
... Visualization of CBs in living plant cells revealed also that plant CBs exhibit dynamic movement, fusing together and splitting apart within the nucleus (Boudonck et al. 1999). Moreover, the number of CBs in plant cells was observed to change during the cell cycle and differentiation (Boudonck et al. 1998; Straatman and Schel 2001; Seguí-Simarro et al. 2006; Zienkiewicz and Bednarska 2009). Recently, plant CBs were shown to contain also Atcoilin (Collier et al. 2006; Koroleva et al. 2009), which is a homolog of the mammalian protein coilin, a marker of CB in animal cells. ...
In microsporocytes of the European larch, we demonstrated the presence of several mRNAs in spherical nuclear bodies. In the nuclei of microsporocytes, we observed up to 12 bodies, ranging from 0.5 to 6 μm in diameter, during the prophase of the first meiotic division. Our previous studies revealed the presence of polyadenylated RNA (poly(A) RNA) in these bodies, but did not confirm the presence of nascent transcripts or splicing factors of the SR family. The lack of these molecules precludes the bodies from being the sites of synthesis and early maturation of primary transcripts (Kołowerzo et al., Protoplasma 236:13–19, 2009). However, the bodies serve as sites for the accumulation of splicing machinery, including the Sm proteins and small nuclear RNAs. Characteristic ultrastructures and the molecular composition of the nuclear bodies, which contain poly(A) RNA, are indicative of Cajal bodies (CBs). Here, we demonstrated the presence of several housekeeping gene transcripts—α-tubulin, pectin methylesterase, peroxidase and catalase, ATPase, and inositol-3-phosphate synthase—in CBs. Additionally, we observed transcripts of the RNA polymerase II subunits RPB2 and RPB10 RNA pol II and the core spliceosome proteins mRNA SmD1, SmD2, and SmE. The co-localization of nascent transcripts and mRNAs indicates that mRNA accumulation/storage, particularly in CBs, occurs in the nucleus of microsporocytes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00412-011-0339-4) contains supplementary material, which is available to authorized users.
... One characteristic trait of Cajal bodies is the dynamic changes in their size and number in different species, individual tissues, and cells (Boudonck et al. 1998; Acevedo et al. 2002). Moreover, it has been shown that the number of CBs in plant cells changes during the cell cycle and differentiation (Boudonck et al. 1998; Straatman and Schel 2001; SeguíSimarro et al. 2006; Zienkiewicz and Bednarska 2009). It has been experimentally established that the number of Cajal bodies is related to the metabolic activity of a cell. ...
... It has been experimentally established that the number of Cajal bodies is related to the metabolic activity of a cell. A positive correlation between the formation of Cajal bodies and transcriptional activity was revealed in HeLa cells (Ferreira et al. 1994), in hamster embryos (Ferreira and Carmo-Fonseca 1996), in oocytes (Chouinard 1975; Parfenov et al. 2003) and during microspore embryogenic development (Simarro et al. 2006), and pollen development (Zienkiewicz and Bednarska 2009) in plant cells. However, there are exceptions. ...
Small nuclear ribonucleoproteins (snRNPs) play a fundamental role in pre-mRNA processing in the nucleus. The biogenesis of snRNPs involves a sequence of events that occurs in both the nucleus and cytoplasm. Despite the wealth of biochemical information about the cytoplasmic assembly of snRNPs, little is known about the spatial organization of snRNPs in the cytoplasm. In the cytoplasm of larch microsporocytes, a cyclic appearance of bodies containing small nuclear RNA (snRNA) and Sm proteins was observed during anther meiosis. We observed a correlation between the occurrence of cytoplasmic snRNP bodies, the levels of Sm proteins, and the dynamic formation of Cajal bodies. Larch microsporocytes were used for these studies. This model is characterized by natural fluctuations in the level of RNA metabolism, in which periods of high transcriptional activity are separated from periods of low transcriptional activity. In designing experiments, the authors considered the differences between the nuclear and cytoplasmic phases of snRNP maturation and generated a hypothesis about the direct participation of Sm proteins in a molecular switch triggering the formation of Cajal bodies.
Electronic supplementary material
The online version of this article (doi:10.1007/s00418-011-0861-8) contains supplementary material, which is available to authorized users.
... In addition to the size difference, the vegetative cell has been shown to be much more active in RNA synthesis than the generative cell (Zienkiewicz et al. 2008). The highest levels of transcription in both cells were observed in young pollen, whereas a successive repression of RNA synthesis was found at later stages of pollen development (Zienkiewicz et al. 2008, Zienkiewicz andBednarska 2009). Microspores and differentiating pollen grains are unique and excellent natural models for RNA synthesis and inhibition of transcription (Zienkiewicz and Bednarska 2009). ...
... The highest levels of transcription in both cells were observed in young pollen, whereas a successive repression of RNA synthesis was found at later stages of pollen development (Zienkiewicz et al. 2008, Zienkiewicz andBednarska 2009). Microspores and differentiating pollen grains are unique and excellent natural models for RNA synthesis and inhibition of transcription (Zienkiewicz and Bednarska 2009). However, little is known about the role of RNA synthesis and the splicing machinery in this system. ...
Flowering plants produce multicellular gametophytes through an elaborate regulation of gametogenesis. During female and male gametogenesis in Arabidopsis thaliana, sporogenous cells differentiate and undergo meiosis to produce megaspores and microspores, which in turn go through mitosis to develop into multicellular gametophytes. Here we report that the Arabidopsis spliceosomal protein, SPLICEOSOME-ASSOCIATED PROTEIN 130 (AtSAP130), is required for proper reproduction. AtSAP130 is encoded by two genes, AtSAP130a and AtSAP130b. Plants with reduced expression of the AtSAP130 genes, induced by RNA interference, showed a defect in fertilization. Besides functional impairment observed in the female reproductive organs, analysis focusing on pollen development revealed defects in the transition from the microspore to the bicellular stage. Our results suggest that AtSAP130a and AtSAP130b play an indispensable role in specific spatiotemporal events in reproduction.
In microsporocytes of the European larch, we demonstrated the presence of several mRNAs in spherical nuclear bodies. In the nuclei of microsporocytes, we observed up to 12 bodies, ranging from 0.5 to 6 μm in diameter, during the prophase of the first meiotic division. Our previous studies revealed the presence of polyadenylated RNA (poly(A) RNA) in these bodies, but did not confirm the presence of nascent transcripts or splicing factors of the SR family. The lack of these molecules precludes the bodies from being the sites of synthesis and early maturation of primary transcripts (Kołowerzo et al., Protoplasma 236:13–19, 2009). However, the bodies serve as sites for the accumulation of splicing machinery, including the Sm proteins and small nuclear RNAs. Characteristic ultrastructures and the molecular composition of the nuclear bodies, which contain poly(A) RNA, are indicative of Cajal bodies (CBs). Here, we demonstrated the presence of several housekeeping gene transcripts—α-tubulin, pectin methylesterase, peroxidase and catalase, ATPase, and inositol-3-phosphate synthase—in CBs. Additionally, we observed transcripts of the RNA polymerase II subunits RPB2 and RPB10 RNA pol II and the core spliceosome proteins mRNA SmD1, SmD2, and SmE. The co-localization of nascent transcripts and mRNAs indicates that mRNA accumulation/storage, particularly in CBs, occurs in the nucleus of microsporocytes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00412-011-0339-4) contains supplementary material, which is available to authorized users.