Figure 1 - available from: BMC Genomics
This content is subject to copyright. Terms and conditions apply.
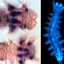
Early development of the marine annelid Platynereis dumerilii.a Dorsal views of Platynereis adults showing the head and anterior segments of a male (top) and female (bottom). b Phylogenetic position of the spiralian annelid Platynereis dumerilii (highlighted in red). Bilaterally symmetric animals comprise of deuterostomes and protostomes. Within protostomes there are two clades, ecdysozoans and lophotrochozoans. Many lophotrochozoan phyla are spiralians. c Early spiral cleavage patterns in Platynereis. The first cleavage is highly asymmetric, giving rise to the AB cell and the much larger CD cell (2-cell). The second cleavage creates the macromeres a, b, c and d (4-cell). The third cleavage (8-cell) is the first cleavage along the animal/vegetal axis and marks the beginning of spiral cleavage. The animal-pole micromeres, denoted with lowercase letters, are shifted slightly clockwise with respect to their macromere sisters, denoted with uppercase letters. At the fourth cleavage (16-cell), animal-pole daughter cells are shifted counter-clockwise compared to their more vegetal sister cells. This spiral pattern of cell divisions continues throughout early embryogenesis and transitions in some cells to bilaterally symmetric cell divisions (66-cell). Cells depicted in green will form multi-ciliated cells by 12 h of development. The four earlier stages show two small polar bodies at the animal pole. d The developmental timeline of Platynereis dumerilii from the zygote to the protrochophore. The first cleavage occurs around 2 h post fertilization (hpf) and the spiral cleavage pattern (described in c) continues throughout the first ~12 h of development. Platynereis transitions to a pattern of bilaterally symmetric cell divisions around 7hpf. The dashed lines show the approximate ages of various cell stages. The time points at which samples were collected for RNA-sequencing are in red
Source publication
Background
The spiral cleavage mode of early development is utilized in over one-third of all animal phyla and generates embryonic cells of different size, position, and fate through a conserved set of stereotypic and invariant asymmetric cell divisions. Despite the widespread use of spiral cleavage, regulatory and molecular features for any spiral...
Citations
... Abundant jelly secretion indicates successful fertilization. During the early stages of P. dumerilii development, embryos exhibit stereotypical spiral cleavage [20]. The initial divisions are highly asymmetric, and cells can be identified based on their position and size. ...
... The second problem was mortality due to water fouling, which mostly resulted from feeding the worms with Sera micron. Initially, we provided quantities of the suspension that were too large for the small number of worms (15)(16)(17)(18)(19)(20)(21)(22)(23)(24)(25)(26)(27)(28)(29)(30) in each box. However, Sera micron, which is primarily made with Spirulina cyanobacteria, is a highly nutritious food and efficient for fast growth of the worms. ...
Platynereis dumerilii , a marine annelid, is a model animal that has gained popularity in various fields such as developmental biology, biological rhythms, nervous system organization and physiology, behaviour, reproductive biology, and epigenetic regulation. The transparency of P . dumerilii tissues at all developmental stages makes it easy to perform live microscopic imaging of all cell types. In addition, the slow-evolving genome of P . dumerilii and its phylogenetic position as a representative of the vast branch of Lophotrochozoans add to its evolutionary significance. Although P . dumerilii is amenable to transgenesis and CRISPR-Cas9 knockouts, its relatively long and indefinite life cycle, as well as its semelparous reproduction have been hindrances to its adoption as a reverse genetics model. To overcome this limitation, an adapted culturing method has been developed allowing much faster life cycling, with median reproductive age at 13–14 weeks instead of 25–35 weeks using the traditional protocol. A low worm density in boxes and a strictly controlled feeding regime are important factors for the rapid growth and health of the worms. This culture method has several advantages, such as being much more compact, not requiring air bubbling or an artificial moonlight regime for synchronized sexual maturation and necessitating only limited water change. A full protocol for worm care and handling is provided.
... As such, at this stage, the regeneration process is finished and posterior growth has resumed [42] (Additional file 1). In terms of genomic and transcriptomic resources, there is currently no published Platynereis dumerilii genome, while some transcriptomic data has been essentially generated from embryonic and larval stages [43][44][45], as well as from brain tissue [46]. Few scRNA-seq datasets from early embryos and larvae have also been generated [47,48]. ...
... The main aim of our study was to investigate the transcriptional landscape of posterior regeneration in an emerging regeneration model: the annelid Platynereis dumerilii [40]. Hence, it was mandatory to reconstruct a high-quality reference transcriptome suitable for the study of this post-embryonic developmental process, as so far available transcriptomes are mostly encompassing early developmental stages or specific tissues [44][45][46]. To this end, we took advantage of a variety of Platynereis' data sources collected from the literature [45,46], and specifically generated additional missing transcriptomic data. ...
... Hence, it was mandatory to reconstruct a high-quality reference transcriptome suitable for the study of this post-embryonic developmental process, as so far available transcriptomes are mostly encompassing early developmental stages or specific tissues [44][45][46]. To this end, we took advantage of a variety of Platynereis' data sources collected from the literature [45,46], and specifically generated additional missing transcriptomic data. ...
Background
Restorative regeneration, the capacity to reform a lost body part following amputation or injury, is an important and still poorly understood process in animals. Annelids, or segmented worms, show amazing regenerative capabilities, and as such are a crucial group to investigate. Elucidating the molecular mechanisms that underpin regeneration in this major group remains a key goal. Among annelids, the nereididae Platynereis dumerilii (re)emerged recently as a front-line regeneration model. Following amputation of its posterior part, Platynereis worms can regenerate both differentiated tissues of their terminal part as well as a growth zone that contains putative stem cells. While this regeneration process follows specific and reproducible stages that have been well characterized, the transcriptomic landscape of these stages remains to be uncovered.
Results
We generated a high-quality de novo Reference transcriptome for the annelid Platynereis dumerilii. We produced and analyzed three RNA-sequencing datasets, encompassing five stages of posterior regeneration, along with blastema stages and non-amputated tissues as controls. We included two of these regeneration RNA-seq datasets, as well as embryonic and tissue-specific datasets from the literature to produce a Reference transcriptome. We used this Reference transcriptome to perform in depth analyzes of RNA-seq data during the course of regeneration to reveal the important dynamics of the gene expression, process with thousands of genes differentially expressed between stages, as well as unique and specific gene expression at each regeneration stage. The study of these genes highlighted the importance of the nervous system at both early and late stages of regeneration, as well as the enrichment of RNA-binding proteins (RBPs) during almost the entire regeneration process.
Conclusions
In this study, we provided a high-quality de novo Reference transcriptome for the annelid Platynereis that is useful for investigating various developmental processes, including regeneration. Our extensive stage-specific transcriptional analysis during the course of posterior regeneration sheds light upon major molecular mechanisms and pathways, and will foster many specific studies in the future.
... As a member of the phylum Annelida, P. dumerilii fills a need for a bilaterian or more specific a spiralian model species in addition to the traditional insect, nematode, and vertebrate models that have dominated much of embryological research. With the recent development of several molecular resources and essential techniques (Altincicek and Vilcinskas, 2007;Chou et al., 2016;Conzelmann et al., 2013;Ozpolat et al., 2021), P. dumerilii is becoming an ideal organism for studying the evolution and conservation of gene functions in bilaterians. As an indirect developing annelid, it goes through a series of embryonic and larval stages that form several ciliated structures that allow for motility in its marine environment (Fig. 1C) (Fischer and Dorresteijn, 2004;Fischer et al., 2010). ...
... All P. dumerilii tektin and foxJ1 sequences were obtained from transcriptomic data (Chou et al., 2016) by tBLASTN and confirmed by reciprocal BLAST and phylogenetic analysis. Primers were designed using the program Primer3 (Rozen and Skaletsky, 1999). ...
... To determine the tektin gene complement of P. dumerilii, tektins were identified from a previously published embryonic transcriptome (Chou et al., 2016) and from an unpublished Platynereis draft genome (Platynereis sequencing consortium & Arendt laboratory) using reciprocal BLAST analysis with annotated H. sapiens sequences as queries. Five P. dumerilii tektins were identified, cloned, sequenced, and confirmed by phylogenetic analysis (Fig. 1B (Bastin and Schneider, 2019)) to encode single copy orthologs for each of the five ancestral spiralian tektin genes. ...
Tektins are a highly conserved family of coiled-coil domain containing proteins known to play a role in structure, stability and function of cilia and flagella. Tektin proteins are thought to form filaments which run the length of the axoneme along the inner surface of the A tubule of each microtubule doublet. Phylogenetic analyses suggest that the tektin family arose via duplications from a single tektin gene in a unicellular organism giving rise to four and five tektin genes in bilaterians and in spiralians, respectively. Although tektins are found in most metazoans, little is known about their expression and function outside of a handful of model species. Here we present the first comprehensive study of tektin family gene expression in any animal system, in the spiralian annelid Platynereis dumerilii. This indirect developing species retains a full ancient spiralian complement of five tektin genes. We show that all five tektins are expressed almost exclusively in known ciliary structures following the expression of the motile cilia master regulator foxJ1. The three older bilaterian tektin-1, tektin-2, and tektin-4 genes, show a high degree of spatial and temporal co-regulation, while the spiralian specific tektin-3/5A and tektin-3/5B show a delay in onset of expression in every ciliary structure. In addition, tektin-3/5B transcripts show a restricted subcellular localization to the most apical region near the multiciliary arrays. The exact recapitulation of the sequence of expression and localization of the five tektins at different times during larval development indicates the cooption of a fixed regulatory and cellular program during the formation of each ciliary band and multiciliated cell type in this spiralian.
... Linking cell lineages and individual cells to gene expression and cell fate is possible in P. dumerilii [57,[60][61][62][63]. Embryos are large enough to microinject for fluorescent labeling of cell nuclei and membrane, they show fast recovery after injection, and are small and transparent enough to image for several days (Additional file 1: Table S1). In addition, various sequencing approaches for obtaining developmental stage-specific transcriptomes have been used [64][65][66]. These resources allow the identification of transcription factors and signaling pathways and assist in mapping gene expression to cell lineages and cell fates. ...
... The discovery of these determinants is now within reach [71,72]. Altogether, these data shed light onto which signaling molecules and pathways are operating during spiralian development, and when and where tissue specification and differentiation take place [63,64,67,[73][74][75] (Fig. 2b''). ...
... Also, gene linkage, or synteny, appears relatively less derived from the urbilaterian condition than in fly and nematode, at least as indicated by the Antennapedia (ANTP)-class homeobox-containing genes, with their largely similar organization relative to chordates [169]. Similarly, P. dumerilii gene sequences appear to be relatively 'short branch' [168] and transcriptomes show a higher similarity of homologues with deuterostomes rather than ecdysozoans [64]. In addition, complements of gene families and composition of pathways show high levels of similarity to what is inferred for the bilaterian ancestor (or even earlier). ...
Abstract
The Nereid Platynereis dumerilii (Audouin and Milne Edwards (Annales des Sciences Naturelles 1:195–269, 1833) is
a marine annelid that belongs to the Nereididae, a family of errant polychaete worms. The Nereid shows a pelagobenthic
life cycle: as a general characteristic for the superphylum of Lophotrochozoa/Spiralia, it has spirally cleaving
embryos developing into swimming trochophore larvae. The larvae then metamorphose into benthic worms living in
self-spun tubes on macroalgae. Platynereis is used as a model for genetics, regeneration, reproduction biology, development,
evolution, chronobiology, neurobiology, ecology, ecotoxicology, and most recently also for connectomics
and single-cell genomics. Research on the Nereid started with studies on eye development and spiralian embryogenesis
in the nineteenth and early twentieth centuries. Transitioning into the molecular era, Platynereis research focused
on posterior growth and regeneration, neuroendocrinology, circadian and lunar cycles, fertilization, and oocyte
maturation. Other work covered segmentation, photoreceptors and other sensory cells, nephridia, and population
dynamics. Most recently, the unique advantages of the Nereid young worm for whole-body volume electron microscopy
and single-cell sequencing became apparent, enabling the tracing of all neurons in its rope-ladder-like central
nervous system, and the construction of multimodal cellular atlases. Here, we provide an overview of current topics
and methodologies for P. dumerilii, with the aim of stimulating further interest into our unique model and expanding
the active and vibrant Platynereis community.
Keywords: Annelida, Spiralia, Marine model species, Evo-devo, Integrative biology
... Detailed and comprehensive analysis of early worm's development transcription dynamics was published in 2016 [20]. In that work seven stages of development were studied, from zygote (2 hpf) to early protrochophore stage of~330 cells (14 hpf). ...
... In that study the oocyte stage was not investigated, so we do not know how comparable their transcription landscapes is. However, one maternal transcript, which we found, Pdum-Hox1, is also described for zygote [20]. Pdum-Hox1 isoform, which was annotated in Chou et al. study [20] as a Hox-B1a, hit in the cluster with majority of other maternal transcripts. ...
... However, one maternal transcript, which we found, Pdum-Hox1, is also described for zygote [20]. Pdum-Hox1 isoform, which was annotated in Chou et al. study [20] as a Hox-B1a, hit in the cluster with majority of other maternal transcripts. Among the zygote maternal transcripts, small amounts of Hox2 and Hox7 mRNA are detected. ...
Hox genes are some of the best studied developmental control genes. In the overwhelming majority of bilateral animals, these genes are sequentially activated along the main body axis during the establishment of the ground plane, i.e., at the moment of gastrulation. Their activation is necessary for the correct differentiation of cell lines, but at the same time it reduces the level of stemness. That is why the chromatin of Hox loci in the pre-gastrulating embryo is in a bivalent state. It carries both repressive and permissive epigenetic markers at H3 histone residues, leading to transcriptional repression. There is a paradox that maternal RNAs, and in some cases the proteins of the Hox genes, are present in oocytes and preimplantation embryos in mammals. Their functions should be different from the zygotic ones and have not been studied to date. Our object is the errant annelid Platynereis dumerilii. This model is convenient for studying new functions and mechanisms of regulation of Hox genes, because it is incomparably simpler than laboratory vertebrates. Using a standard RT-PCR on cDNA template which was obtained by reverse transcription using random primers, we found that maternal transcripts of almost all Hox genes are present in unfertilized oocytes of worm. We assessed the localization of these transcripts using WMISH.
... Determining the CpG observed/expected (o/e) ratios can thus be used to estimate 5mC levels: CpG o/e close to 1 means no methylation while CpG o/e far below 1 suggests that methylation of CpGs is present (e.g., [41][42][43]). As in non-vertebrates methylated CpGs are mostly found within gene bodies [11,12], we calculated CpG o/ e for P. dumerilii gene bodies, by applying Notos, a software that computes CpG o/e ratios based on kernel density estimations [43,48], on a high-quality P. dumerilii reference transcriptome [49]. We found a CpG o/e distribution with a single mode at 0.55 (Fig. 1a), suggesting high-level gene body methylation in P. dumerilii. ...
... We first took advantage of two previously published transcriptomic datasets corresponding to various developmental stages and adult conditions of P. dumerilii, available in a public database (PdumBase) [59]. The first dataset corresponds to embryonic developmental stages, ranging from 2 to 14hpf, with a time point every 2 h [49]. The second dataset comprises major larval stages (24 to 4dpf; five time points), juvenile stages (10dpf to 3mpf; five time points), and adult reproductive stages (males and females) [60]. ...
... Expression values for most genes (exceptions are Pdum-dnmt3 absent in the two sets of transcriptomic data and Pdum-gatad2 and Pdum-rbbp4/7 only found as chimeric transcripts) were recovered and can be found in Additional file 7: Fig. S5. High transcript levels are found for many studied genes in the earliest developmental stages (2-6hpf) and several of them belong to co-expression clusters defined by Chou et al. [49] as maternal gene clusters (clusters 1-4; Additional file 7: Fig. S5). This indicates that the P. dumerilii egg contains a large pool of maternal transcripts coding for DNA methylation proteins that could be used for embryonic development. ...
Background
Methylation of cytosines in DNA (5mC methylation) is a major epigenetic modification that modulates gene expression and constitutes the basis for mechanisms regulating multiple aspects of embryonic development and cell reprogramming in vertebrates. In mammals, 5mC methylation of promoter regions is linked to transcriptional repression. Transcription regulation by 5mC methylation notably involves the nucleosome remodeling and deacetylase complex (NuRD complex) which bridges DNA methylation and histone modifications. However, less is known about regulatory mechanisms involving 5mC methylation and their function in non-vertebrate animals. In this paper, we study 5mC methylation in the marine annelid worm Platynereis dumerilii, an emerging evolutionary and developmental biology model capable of regenerating the posterior part of its body post-amputation.
Results
Using in silico and experimental approaches, we show that P. dumerilii displays a high level of DNA methylation comparable to that of mammalian somatic cells. 5mC methylation in P. dumerilii is dynamic along the life cycle of the animal and markedly decreases at the transition between larval to post-larval stages. We identify a full repertoire of mainly single-copy genes encoding the machinery associated with 5mC methylation or members of the NuRD complex in P. dumerilii and show that this repertoire is close to the one inferred for the last common ancestor of bilaterians. These genes are dynamically expressed during P. dumerilii development and regeneration. Treatment with the DNA hypomethylating agent Decitabine impairs P. dumerilii larval development and regeneration and has long-term effects on post-regenerative growth.
Conclusions
Our data reveal high levels of 5mC methylation in the annelid P. dumerilii, highlighting that this feature is not specific to vertebrates in the bilaterian clade. Analysis of DNA methylation levels and machinery gene expression during development and regeneration, as well as the use of a chemical inhibitor of DNA methylation, suggest an involvement of 5mC methylation in P. dumerilii development and regeneration. We also present data indicating that P. dumerilii constitutes a promising model to study biological roles and mechanisms of DNA methylation in non-vertebrate bilaterians and to provide new knowledge about evolution of the functions of this key epigenetic modification in bilaterian animals.
... Preliminary data revealed that Platynereis genome appears less compact than in other annelids (~1 to 2 Gpb) (Zantke et al. 2014), and a previous analysis comparing bacterial artificial chromosome sequencing (BAC) and expressed sequence tags (EST) on a subset of 30 randomly-detected genes suggested that Platynereis genes are intron-rich, surprisingly with two-thirds of introns shared between Platynereis and human orthologs . Various additional transcriptomic databases have been acquired during the past years (Table 13.1), including bulk RNA-seq data for all key stages of embryonic and larval development, juveniles of different ages, and adults (Conzelmann et al. 2013;Chou et al. 2016). These data have been grouped altogether and are now publicly available on a website Pdumbase platform (Chou et al. 2018) (pdumbase.gdcb.iastate.edu). ...
... Determining the CpG observed/expected (o/e) ratios can thus be used to estimate 5mC levels: CpG o/e close to 1 means no methylation while CpG o/e far below 1 suggests that methylation of CpGs is present (e.g., [41][42][43]). As in non-vertebrates methylated CpGs are mostly found within gene bodies [11,12], we calculated CpG o/ e for P. dumerilii gene bodies, by applying Notos, a software that computes CpG o/e ratios based on kernel density estimations [43,48], on a high-quality P. dumerilii reference transcriptome [49]. We found a CpG o/e distribution with a single mode at 0.55 (Fig. 1a), suggesting high-level gene body methylation in P. dumerilii. ...
... We first took advantage of two previously published transcriptomic datasets corresponding to various developmental stages and adult conditions of P. dumerilii, available in a public database (PdumBase) [59]. The first dataset corresponds to embryonic developmental stages, ranging from 2 to 14hpf, with a time point every 2 h [49]. The second dataset comprises major larval stages (24 to 4dpf; five time points), juvenile stages (10dpf to 3mpf; five time points), and adult reproductive stages (males and females) [60]. ...
... Expression values for most genes (exceptions are Pdum-dnmt3 absent in the two sets of transcriptomic data and Pdum-gatad2 and Pdum-rbbp4/7 only found as chimeric transcripts) were recovered and can be found in Additional file 7: Fig. S5. High transcript levels are found for many studied genes in the earliest developmental stages (2-6hpf) and several of them belong to co-expression clusters defined by Chou et al. [49] as maternal gene clusters (clusters 1-4; Additional file 7: Fig. S5). This indicates that the P. dumerilii egg contains a large pool of maternal transcripts coding for DNA methylation proteins that could be used for embryonic development. ...
... Determination of CpG observed/expected (o/e) ratio can thus be used as an estimator of 5mC levels, CpG o/e close to 1 means no methylation while CpG o/e far below 1 suggests that methylation of CpGs is present (e.g., [38][39][40]). As in non-vertebrates methylated CpGs are mostly found within gene bodies [10,11], we calculated CpG o/e for P. dumerilii gene bodies, by applying Notos, a software that computes CpG o/e ratios based on kernel density estimations [40,44], on a high quality P. dumerilii reference transcriptome [45]. We found a CpG o/e distribution with a single mode at 0.55 ( Figure 1A), suggesting high-level gene body methylation in P. dumerilii. ...
... dumerilii. We first took advantage of two previously published transcriptomic datasets corresponding to various developmental stages and adult condition of P. dumerilii [45,54], available in a public database (Pdumbase) [55]. The first dataset corresponds to embryonic developmental stages, ranging from 2 to 14hhpf, with a time point every two hours [45]. ...
... We first took advantage of two previously published transcriptomic datasets corresponding to various developmental stages and adult condition of P. dumerilii [45,54], available in a public database (Pdumbase) [55]. The first dataset corresponds to embryonic developmental stages, ranging from 2 to 14hhpf, with a time point every two hours [45]. The second dataset comprises major larval stages (24hpf to 4dpf; five time points), juvenile stages (10dpf to 3 months post fertilization (mpf); five time points) and adult reproductive stages (males and females) [54]. ...
Background
Methylation of cytosines in DNA (5mC methylation) is a major epigenetic modification that modulates gene expression and is important for embryonic development and cell reprogramming in vertebrates. In mammals, 5mC methylation in promoter regions is linked to transcriptional repression. Transcription regulation by 5mC methylation notably involves the Nucleosome Remodeling and Deacetylase complex (NuRD complex) which bridges DNA methylation and histone modifications. Less is known about roles and mechanisms of 5mC methylation in non-vertebrate animals. In this paper, we study 5mC methylation in the marine annelid worm Platynereis dumerilii, an emerging evolutionary and developmental biology model capable of regenerating the posterior part of its body upon amputation. The regenerated region includes both differentiated structures and a growth zone consisting of stem cells required for the continuous growth of the worm.
Results
Using in silico and experimental approaches, we show that P. dumerilii displays a high level of DNA methylation comparable to that of mammalian somatic cells. 5mC methylation in P. dumerilii is dynamic along the life cycle of the animal and markedly decreases at the transition between larval to post-larval stages. We identify a full repertoire of mainly singlecopy genes encoding the machinery associated to 5mC methylation or members of the NuRD complex in P. dumerilii and show, through phylogenetic analyses, that this repertoire is close to the one inferred for the last common ancestor of bilaterians. These genes are dynamically expressed during P. dumerilii development, growth and regeneration. Treatment with the DNA hypomethylating agent Decitabine, impairs P. dumerilii larval development and regeneration, and has long-term effects on post-regenerative growth by affecting the functionality of stem cells of the growth zone.
Conclusions
Our data indicate high-level of 5mC methylation in the annelid P. dumerilii, highlighting that this feature is not specific to vertebrates in the bilaterian clade. Analysis of DNA methylation levels and machinery gene expression during development and regeneration, as well as the use of a chemical inhibitor of DNA methylation, suggest an involvement of 5mC methylation in P. dumerilii development, regeneration and stem cell-based post-regenerative growth. We also present data indicating that P. dumerilii constitutes a promising model to study biological roles and mechanisms of DNA methylation in non-vertebrate bilaterians and to provide new knowledge about evolution of the functions of this key epigenetic modification in bilaterian animals.
... Platynereis dumerilii embryos were obtained from a culture at Iowa State University, and originated from a laboratory strain from the University of Mainz, Germany 62 . Lophotrochin and trochin sequences were obtained by tBLASTn searches of transcriptomic data 63 . Genes were cloned by PCR amplification with GoTaq (Promega) using gene specific primers for each gene. ...
Spiralia is a large, ancient and diverse clade of animals, with a conserved early developmental program but diverse larval and adult morphologies. One trait shared by many spiralians is the presence of ciliary bands used for locomotion and feeding. To learn more about spiralian-specific traits we have examined the expression of 20 genes with protein motifs that are strongly conserved within the Spiralia, but not detectable outside of it. Here, we show that two of these are specifically expressed in the main ciliary band of the mollusc Tritia (also known as Ilyanassa). Their expression patterns in representative species from five more spiralian phyla—the annelids, nemerteans, phoronids, brachiopods and rotifers—show that at least one of these, lophotrochin, has a conserved and specific role in particular ciliated structures, most consistently in ciliary bands. These results highlight the potential importance of lineage-specific genes or protein motifs for understanding traits shared across ancient lineages. Spiralians have ciliary bands, used for locomotion and feeding, but defining molecular features of these structures are unknown. Here, the authors report a gene, Lophotrochin, that contains a protein domain only found in spiralians, and specifically expressed in diverse ciliary bands across the group, which provides a molecular signature for these structures.