Fig 3 - uploaded by Alfredo De Bustos
Content may be subject to copyright.
Source publication
The products of the Nbs1, Mre11, and Rad50 genes form the MRN complex. This complex is involved mainly in DNA repair, but it also has roles in meiosis, replication, telomere maintenance, and cell cycle checkpoint signaling. Its study is therefore of great importance if our knowledge of these important cell processes is to improve. To date, few stud...
Contexts in source publication
Context 1
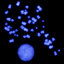
... and 5SrDNA signals were mapped to different arms of chromosome 1D ( fig. 3a). In Triticum aestivum, three loci, Nbs1A, Nbs1B, and Nbs1D, none of which have been previ- ously mapped, were localized to similar positions on the long arm of chromosomes 1A, 1B, and 1D, respectively ( fig. ...
Context 2
... and 5SrDNA signals were mapped to different arms of chromosome 1D ( fig. 3a). In Triticum aestivum, three loci, Nbs1A, Nbs1B, and Nbs1D, none of which have been previ- ously mapped, were localized to similar positions on the long arm of chromosomes 1A, 1B, and 1D, respectively ( fig. ...
Context 3
... alignment of deduced proteins for the characterized Nbs1 genes (see fig. A3 in the online edition of the Interna- tional Journal of Plant Sciences) highlighted their strong de- gree of conservation (98%-99% homology). To compare the homology of wheat proteins with previously characterized se- quences, a phylogenetic tree was constructed ( fig. 4) from the alignment of the deduced proteins with those of rice, ...
Context 4
... alignment of the proteins also showed that wheat NBS1 contains the most important motifs and domains de- scribed in other NBS1 proteins (see fig. 5 for a diagram of the gene; further information is provided in fig. A3). Thus, the predicted FHA and BRTC1 domains of wheat proteins perfectly match those described for rice and maize ( Akutsu et al. 2007;Waterworth et al. 2007). A putative BRTC2 do- main was also identified, based on the a/b topology recorded when using the services of the PredictProtein Server. Wheat NBS1 proteins showed the highly ...
Similar publications
The non-random three-dimensional (3D) organization of the genome in the nucleus is critical to gene regulation and genome function. Using high-throughput chromatin conformation capture, we generated chromatin interaction maps for Brassica rapa and Brassica oleracea at a high resolution and characterized the conservation and divergence of chromatin...
Citations
... Moreover, the MMR and HR pathways can be mutually dependent, and MMR proteins are involved in HR [33,38,39]. Homoeologous genes of the MRN complex, including MRE11, are expressed at different levels in different polyploid wheat species and different tissues-specifically, roots, leaves, and meiocytes [40]. As a complement to this finding, our data indicate the influence of abiotic factors in the wild population on the expression level of MRE11. ...
A complex DNA repair network maintains genome integrity and genetic stability. In this study, the influence of edaphic factors on DNA damage and repair in wild wheat Triticum dicoccoides was addressed. Plants inhabiting two abutting microsites with dry terra rossa and humid basalt soils were studied. The relative expression level of seven genes involved in DNA repair pathways—RAD51, BRCA1, LigIV, KU70, MLH1, MSH2, and MRE11—was assessed using quantitative real-time PCR (qPCR). Immunolocalization of RAD51, LigIV, γH2AX, RNA Polymerase II, and DNA-RNA hybrid [S9.6] (R-loops) in somatic interphase nuclei and metaphase chromosomes was carried out in parallel. The results showed a lower expression level of genes involved in DNA repair and a higher number of DNA double-strand breaks (DSBs) in interphase nuclei in plants growing in terra rossa soil compared with plants in basalt soil. Further, the number of DSBs and R-loops in metaphase chromosomes was also greater in plants growing on terra rossa soil. Finally, RAD51 and LigIV foci on chromosomes indicate ongoing DSB repair during the M-phase via the Homologous Recombination and Non-Homologous End Joining pathways. Together, these results show the impact of edaphic factors on DNA damage and repair in the wheat genome adapted to contrasting environments.
... T. turgidum itself is derived from a previous hybridisation event that happened some 0.5-3 million years ago between T. urartu (the diploid donor of the A genome) and an unknown B genome donor species (probably Ae. speltoides or a related species). Our group works routinely with species of different ploidy level belonging to the genera Triticum and Aegilops [26][27][28], but reliable comparisons of gene expression have not been possible given the cited problem in the use of the reference genes. To date, few publications exist on interspecific expression analysis in polyploid species, and in no case have reference genes been proposed to normalize qPCR data with respect to ploidy. ...
... cDNA synthesis First strand cDNA synthesis was performed using 1 lg of DNA-free RNA using the Transcriptor Reverse Transcriptase and Oligo (dT) 15 primer (Roche) in a 20 ll total reaction volume. The absence of contaminant DNA was verified by PCR using primers Nbs Seq-L2/Nbs Seq-R2 [28] designed in introns. Success in RT reactions was controlled by PCR with primers qNbs-L3/Nbs Seq-R [28] to amplify control cDNA. ...
... The absence of contaminant DNA was verified by PCR using primers Nbs Seq-L2/Nbs Seq-R2 [28] designed in introns. Success in RT reactions was controlled by PCR with primers qNbs-L3/Nbs Seq-R [28] to amplify control cDNA. For qPCR reactions, 1:4 dilutions were prepared from the 20 ll solutions of cDNA; 1 ll was used for each reaction (equivalent to 12.5 ng of input RNA). ...
Interspecific comparative studies require that expression data be comparable among species, and when species with different levels of ploidy are contemplated the relative expression per cell should be obtained for accurate comparisons to be made. Quantitative reverse-transcription-PCR is the most popular and sensitive technique for the detection and quantification of mRNA in gene expression analysis. In recent years it has become clear that the choice of reference genes for the normalization of expression data is very important. Several studies have shown that the expression of the traditional housekeeping genes varies under certain situations; their use as reference genes in quantitative PCR assays can therefore lead to errors when interpreting the relative expression of target genes. Normalizing with respect to endogenous genes showing a constant level of expression per cell across species, however, provides an easy way of obtaining comparable expression data for other genes in those species. In this work, the validity of several candidate genes was examined across four diploid and polyploid species of the genera Triticum and Aegilops. Candidate reference genes were chosen among the traditional housekeeping genes used in quantitative PCR analysis, as well as others found to have stable levels of expression under different conditions in other studies. After the analyses, candidate genes were gathered into two groups according to the different levels of expression per cell seen in polyploid species. For the four species studied, two genes suitable for normalization procedures in interspecific studies were identified: cell division control protein and malate dehydrogenase. Both showed a constant number of transcripts per cell, independent of the level of ploidy.
Polyploidy is a prominent route to speciation in plants; however, this entails resolving the challenges of meiotic instability facing abrupt doubling of chromosome complement. This issue remains poorly understood.
We subjected progenies of a synthetic hexaploid wheat, analogous to natural common wheat, but exhibiting extensive meiotic chromosome instability, to heat or salt stress. We selected stress‐tolerant cohorts and generated their progenies under normal condition. We conducted fluorescent in situ hybridization/genomic in situ hybridization‐based meiotic/mitotic analysis, RNA ‐Seq and virus‐induced gene silencing ( VIGS )‐mediated assay of meiosis candidate genes.
We show that heritability of stress tolerance concurred with increased euploidy frequency due to enhanced meiosis stability. We identified a set of candidate meiosis genes with altered expression in the stress‐tolerant plants vs control, but the expression was similar to that of common wheat (cv Chinese Spring, CS ). We demonstrate VIGS ‐mediated downregulation of individual candidate meiosis genes in CS is sufficient to confer an unstable meiosis phenotype mimicking the synthetic wheat.
Our results suggest that heritable regulatory changes of preexisting meiosis genes may be hitchhiked as a spandrel of stress tolerance, which significantly improves meiosis stability in the synthetic wheat. Our findings implicate a plausible scenario that the meiosis machinery in hexaploid wheat may have already started to evolve at its onset stage.
The Mre11-Rad50-Nbs1 (MRN) complex is providing paradigm-shifting results of exceptional biomedical interest. MRN is among the earliest respondents to DNA double-strand breaks (DSBs), and MRN mutations cause the human cancer predisposition diseases Nijmegen breakage syndrome and ataxia telangiectasia-like disorder (ATLD). MRN's 3-protein multidomain composition promotes its central architectural, structural, enzymatic, sensing, and signaling functions in DSB responses. To organize the MRN complex, the Mre11 exonuclease directly binds Nbs1, DNA, and Rad50. Rad50, a structural maintenance of chromosome (SMC) related protein, employs its ATP-binding cassette (ABC) ATPase, Zn hook, and coiled coils to bridge DSBs and facilitate DNA end processing by Mre11. Contributing to MRN regulatory roles, Nbs1 harbors N-terminal phosphopeptide interacting FHA and BRCT domains, as well as C-terminal ataxia telangiectasia mutated (ATM) kinase and Mre11 interaction domains. Current emerging structural and biological evidence suggests that MRN has 3 coupled critical roles in DSB sensing, stabilization, signaling, and effector scaffolding: (1) expeditious establishment of protein--nucleic acid tethering scaffolds for the recognition and stabilization of DSBs; (2) initiation of DSB sensing, cell-cycle checkpoint signaling cascades, and establishment of epigenetic marks via the ATM kinase; and (3) functional regulation of chromatin remodeling in the vicinity of a DSB.